Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1
- PMID: 29379020
- PMCID: PMC5789076
- DOI: 10.1038/s41467-017-02770-z
Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1
Abstract
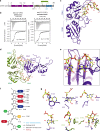
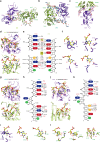
Human hnRNP A2/B1 is an RNA-binding protein that plays important roles in many biological processes, including maturation, transport, and metabolism of mRNA, and gene regulation of long noncoding RNAs. hnRNP A2/B1 was reported to control the microRNAs sorting to exosomes and promote primary microRNA processing as a potential m6A "reader." hnRNP A2/B1 contains two RNA recognition motifs that provide sequence-specific recognition of RNA substrates. Here, we determine crystal structures of tandem RRM domains of hnRNP A2/B1 in complex with various RNA substrates, elucidating specific recognitions of AGG and UAG motifs by RRM1 and RRM2 domains, respectively. Further structural and biochemical results demonstrate multivariant binding modes for sequence-diversified RNA substrates, supporting a RNA matchmaker mechanism in hnRNP A2/B1 function. Moreover, our studies in combination with bioinformatic analysis suggest that hnRNP A2/B1 may mediate effects of m6A through a "m6A switch" mechanism, instead of acting as a direct "reader" of m6A modification.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

