COX16 promotes COX2 metallation and assembly during respiratory complex IV biogenesis
- PMID: 29381136
- PMCID: PMC5809144
- DOI: 10.7554/eLife.32572
COX16 promotes COX2 metallation and assembly during respiratory complex IV biogenesis
Abstract
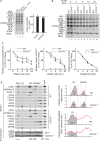
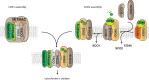
Cytochrome c oxidase of the mitochondrial oxidative phosphorylation system reduces molecular oxygen with redox equivalent-derived electrons. The conserved mitochondrial-encoded COX1- and COX2-subunits are the heme- and copper-center containing core subunits that catalyze water formation. COX1 and COX2 initially follow independent biogenesis pathways creating assembly modules with subunit-specific, chaperone-like assembly factors that assist in redox centers formation. Here, we find that COX16, a protein required for cytochrome c oxidase assembly, interacts specifically with newly synthesized COX2 and its copper center-forming metallochaperones SCO1, SCO2, and COA6. The recruitment of SCO1 to the COX2-module is COX16- dependent and patient-mimicking mutations in SCO1 affect interaction with COX16. These findings implicate COX16 in CuA-site formation. Surprisingly, COX16 is also found in COX1-containing assembly intermediates and COX2 recruitment to COX1. We conclude that COX16 participates in merging the COX1 and COX2 assembly lines.
Keywords: Copper chaperone; Cytochrome c oxidase; Mitochondrial oxidative phosphorylation; biochemistry; cell biology; human; mitochondria; mitochondrial diseases; protein assembly.
© 2018, Aich et al.
Conflict of interest statement
AA, CW, AC, CR, DP, RR, SD, PR No competing interests declared
Figures




References
-
- Baertling F, A M van den Brand M, Hertecant JL, Al-Shamsi A, P van den Heuvel L, Distelmaier F, Mayatepek E, Smeitink JA, Nijtmans LG, Rodenburg RJ. Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Human Mutation. 2015;36:34–38. doi: 10.1002/humu.22715. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

