Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer
- PMID: 29382334
- PMCID: PMC5791351
- DOI: 10.1186/s12943-018-0767-3
Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer
Abstract
Background: Emerging evidence suggests that PIWI-interacting RNAs (piRNAs) may be important epigenetic regulators of gene expression in human cancers; however, their functional and clinical significance in colorectal cancer (CRC) remains unknown.
Methods: We performed piRNA expression profiling in paired cancer and normal tissues through small RNA-sequencing. The clinical significance of candidate piRNAs was investigated, and independently validated in 771 CRC patients from three independent cohorts. The biological function of piRNAs was characterized in cell lines, followed by identification and validation of downstream target genes in CRC tissues.
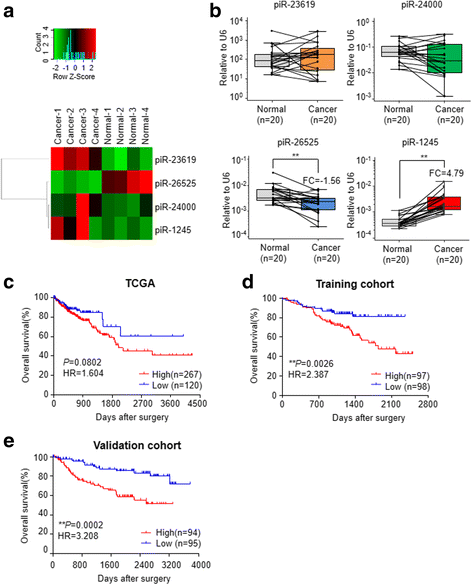
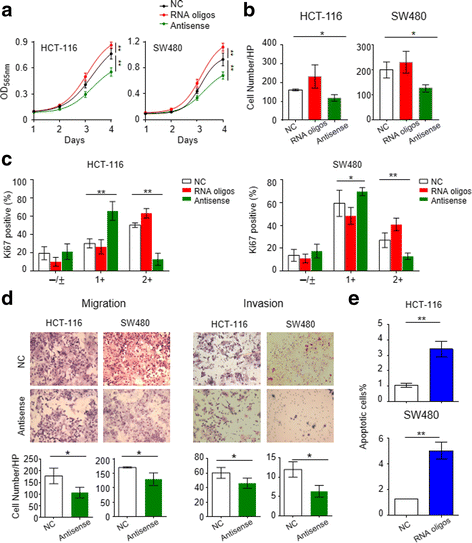
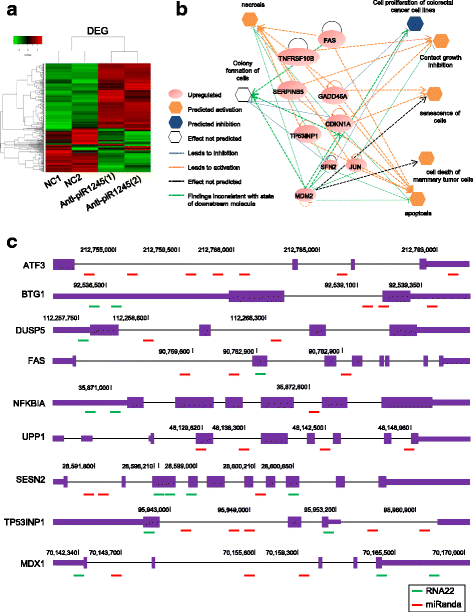
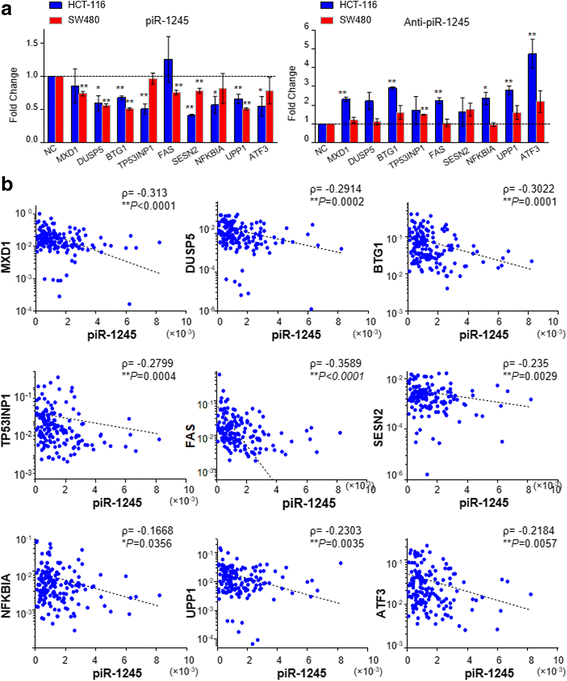
Results: We identified piR-1245 as a novel and frequently overexpressed noncoding RNA in CRC, and its expression significantly correlated with advanced and metastatic disease. Patients with high piR-1245 expression experienced significantly shorter overall survival, and multivariate analysis identified its expression to serve as an independent prognostic biomarker in CRC. Functionally, piR-1245 acts as an oncogene and promotes tumor progression, and gene expression profiling results identified a panel of downstream target-genes involved in regulating cell survival pathway. Based upon piRNA:mRNA sequence complementarity, we identified a panel of tumor suppressor genes (ATF3, BTG1, DUSP1, FAS,NFKBIA, UPP1, SESN2, TP53INP1 and MDX1) as direct targets of piR-1245, and successfully validated an inverse correlation between their expression and piR-1245 in CRC.
Conclusions: We for the first time have identified the role for a PIWI-interacting noncoding RNA, piR-1245, as a novel oncogene and a potential prognostic biomarker in colorectal cancer.
Keywords: Biomarker; Colorectal cancer; Noncoding RNA; Oncogene; Prognosis; Tumor suppressor; piR-1245; piRNA.
Conflict of interest statement
Ethics approval and consent to participate
Written informed consent was obtained from all patients, and the study was approved by the institutional review boards of all participating institutions (Shanghai Tenth People’s Hospital and Okayama University Medical Hospital).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Serum PIWI-Interacting RNAs piR-020619 and piR-020450 Are Promising Novel Biomarkers for Early Detection of Colorectal Cancer.Cancer Epidemiol Biomarkers Prev. 2020 May;29(5):990-998. doi: 10.1158/1055-9965.EPI-19-1148. Epub 2020 Feb 17. Cancer Epidemiol Biomarkers Prev. 2020. PMID: 32066615
-
Small RNA Profiling of piRNAs in Colorectal Cancer Identifies Consistent Overexpression of piR-24000 That Correlates Clinically with an Aggressive Disease Phenotype.Cancers (Basel). 2020 Jan 12;12(1):188. doi: 10.3390/cancers12010188. Cancers (Basel). 2020. PMID: 31940941 Free PMC article.
-
piRNA-823 Is a Unique Potential Diagnostic Non-Invasive Biomarker in Colorectal Cancer Patients.Genes (Basel). 2021 Apr 19;12(4):598. doi: 10.3390/genes12040598. Genes (Basel). 2021. PMID: 33921704 Free PMC article.
-
The emerging role of the piRNA/piwi complex in cancer.Mol Cancer. 2019 Aug 9;18(1):123. doi: 10.1186/s12943-019-1052-9. Mol Cancer. 2019. PMID: 31399034 Free PMC article. Review.
-
The function of novel small non-coding RNAs (piRNAs, tRFs) and PIWI protein in colorectal cancer.Cancer Treat Res Commun. 2022;31:100542. doi: 10.1016/j.ctarc.2022.100542. Epub 2022 Mar 1. Cancer Treat Res Commun. 2022. PMID: 35248886 Review.
Cited by
-
Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications.Biochim Biophys Acta Rev Cancer. 2019 Jan;1871(1):160-169. doi: 10.1016/j.bbcan.2018.12.005. Epub 2018 Dec 30. Biochim Biophys Acta Rev Cancer. 2019. PMID: 30599187 Free PMC article. Review.
-
PIWIL1 interacting RNA piR-017724 inhibits proliferation, invasion, and migration, and inhibits the development of HCC by silencing PLIN3.Front Oncol. 2023 Jul 11;13:1203821. doi: 10.3389/fonc.2023.1203821. eCollection 2023. Front Oncol. 2023. PMID: 37503320 Free PMC article.
-
The Biogenesis and Functions of piRNAs in Human Diseases.Mol Ther Nucleic Acids. 2020 Sep 4;21:108-120. doi: 10.1016/j.omtn.2020.05.023. Epub 2020 May 23. Mol Ther Nucleic Acids. 2020. PMID: 32516734 Free PMC article. Review.
-
Identification and characterization of Piwi-interacting RNAs in human placentas of preeclampsia.Sci Rep. 2021 Aug 3;11(1):15766. doi: 10.1038/s41598-021-95307-w. Sci Rep. 2021. PMID: 34344990 Free PMC article.
-
Integration of multi-omics data to unveil the molecular landscape and role of piRNAs in early-onset colorectal cancer.BMC Med. 2025 Apr 29;23(1):250. doi: 10.1186/s12916-025-04074-2. BMC Med. 2025. PMID: 40301858 Free PMC article.
References
-
- Wilkes GM. Metastatic colorectal cancer: management challenges and opportunities. Oncology (Williston Park) 2011;25(7 Suppl Nurse Ed):32–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

