Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function
- PMID: 29382771
- PMCID: PMC5868693
- DOI: 10.1105/tpc.17.00579
Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function
Abstract
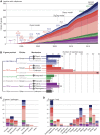
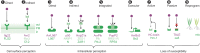
Plants have many, highly variable resistance (R) gene loci, which provide resistance to a variety of pathogens. The first R gene to be cloned, maize (Zea mays) Hm1, was published over 25 years ago, and since then, many different R genes have been identified and isolated. The encoded proteins have provided clues to the diverse molecular mechanisms underlying immunity. Here, we present a meta-analysis of 314 cloned R genes. The majority of R genes encode cell surface or intracellular receptors, and we distinguish nine molecular mechanisms by which R proteins can elevate or trigger disease resistance: direct (1) or indirect (2) perception of pathogen-derived molecules on the cell surface by receptor-like proteins and receptor-like kinases; direct (3) or indirect (4) intracellular detection of pathogen-derived molecules by nucleotide binding, leucine-rich repeat receptors, or detection through integrated domains (5); perception of transcription activator-like effectors through activation of executor genes (6); and active (7), passive (8), or host reprogramming-mediated (9) loss of susceptibility. Although the molecular mechanisms underlying the functions of R genes are only understood for a small proportion of known R genes, a clearer understanding of mechanisms is emerging and will be crucial for rational engineering and deployment of novel R genes.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures
References
-
- Albert I., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1: 15140. - PubMed
-
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

