Ubiquitin C decrement plays a pivotal role in replicative senescence of bone marrow mesenchymal stromal cells
- PMID: 29382826
- PMCID: PMC5833785
- DOI: 10.1038/s41419-017-0032-5
Ubiquitin C decrement plays a pivotal role in replicative senescence of bone marrow mesenchymal stromal cells
Abstract
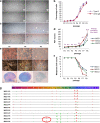
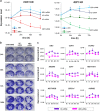
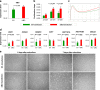
Human bone marrow-mesenchymal stromal cells (hBM-MSCs) undergo cellular senescence during in vitro culture. In this study, we defined this replicative senescence as impaired proliferation, deterioration in representative cell characteristics, accumulated DNA damage, and decreased telomere length and telomerase activity with or without genomic abnormalities. The UBC gene expression gradually decreased during passaging along with the reduction in series of molecules including hub genes; CDK1, CCNA2, MCM10, E2F1, BRCA1, HIST1H1A and HIST1H3B. UBC knockdown in hBM-MSCs induced impaired proliferation in dose-dependent manner and showed replicative senescence-like phenomenon. Gene expression changes after UBC knockdown were similar to late passage hBM-MSCs. Additionally, UBC overexpession improved the proliferation activity of hBM-MSCs accompanied by increased expression of the hub genes. Consequently, UBC worked in higher-order through regulation of the hub genes controlling cell cycle and proliferation. These results indicate that the decrement of UBC expression plays a pivotal role in replicative senescence of hBM-MSCs.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

