Single-cell replication profiling to measure stochastic variation in mammalian replication timing
- PMID: 29382831
- PMCID: PMC5789892
- DOI: 10.1038/s41467-017-02800-w
Single-cell replication profiling to measure stochastic variation in mammalian replication timing
Abstract
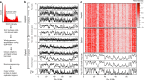
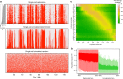
Mammalian DNA replication is regulated via multi-replicon segments that replicate in a defined temporal order during S-phase. Further, early/late replication of RDs corresponds to active/inactive chromatin interaction compartments. Although replication origins are selected stochastically, variation in replication timing is poorly understood. Here we devise a strategy to measure variation in replication timing using DNA copy number in single mouse embryonic stem cells. We find that borders between replicated and unreplicated DNA are highly conserved between cells, demarcating active and inactive compartments of the nucleus. Fifty percent of replication events deviated from their average replication time by ± 15% of S phase. This degree of variation is similar between cells, between homologs within cells and between all domains genomewide, regardless of their replication timing. These results demonstrate that stochastic variation in replication timing is independent of elements that dictate timing or extrinsic environmental variation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

