HTLV-1 Tax plugs and freezes UPF1 helicase leading to nonsense-mediated mRNA decay inhibition
- PMID: 29382845
- PMCID: PMC5789848
- DOI: 10.1038/s41467-017-02793-6
HTLV-1 Tax plugs and freezes UPF1 helicase leading to nonsense-mediated mRNA decay inhibition
Abstract
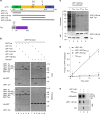
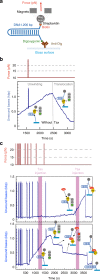
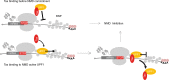
Up-Frameshift Suppressor 1 Homolog (UPF1) is a key factor for nonsense-mediated mRNA decay (NMD), a cellular process that can actively degrade mRNAs. Here, we study NMD inhibition during infection by human T-cell lymphotropic virus type I (HTLV-1) and characterise the influence of the retroviral Tax factor on UPF1 activity. Tax interacts with the central helicase core domain of UPF1 and might plug the RNA channel of UPF1, reducing its affinity for nucleic acids. Furthermore, using a single-molecule approach, we show that the sequential interaction of Tax with a RNA-bound UPF1 freezes UPF1: this latter is less sensitive to the presence of ATP and shows translocation defects, highlighting the importance of this feature for NMD. These mechanistic insights reveal how HTLV-1 hijacks the central component of NMD to ensure expression of its own genome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

