Molecular epidemiology and comparative genomics of Campylobacter concisus strains from saliva, faeces and gut mucosal biopsies in inflammatory bowel disease
- PMID: 29382867
- PMCID: PMC5790007
- DOI: 10.1038/s41598-018-20135-4
Molecular epidemiology and comparative genomics of Campylobacter concisus strains from saliva, faeces and gut mucosal biopsies in inflammatory bowel disease
Abstract
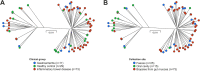
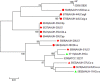
Campylobacter concisus is an emerging pathogen associated with inflammatory bowel disease (IBD), yet little is known about the genetic diversity of C. concisus in relation to host niches and disease. We isolated 104 C. concisus isolates from saliva, mucosal biopsies and faecal samples from 41 individuals (26 IBD, 3 Gastroenteritis (GE), 12 Healthy controls (HC)). Whole genomes were sequenced and the dataset pan-genome examined, and genomic information was used for typing using multi-locus-sequence typing (MLST). C. concisus isolates clustered into two main groups/genomospecies (GS) with 71 distinct sequence types (STs) represented. Sampling site (p < 0.001), rather than disease phenotype (p = 1.00) was associated with particular GS. We identified 97 candidate genes associated with increase or decrease in prevalence during the anatomical descent from the oral cavity to mucosal biopsies to faeces. Genes related to cell wall/membrane biogenesis were more common in oral isolates, whereas genes involved in cell transport, metabolism and secretory pathways were more prevalent in enteric isolates. Furthermore, there was no correlation between individual genetic diversity and clinical phenotype. This study confirms the genetic heterogeneity of C. concisus and provides evidence that genomic variation is related to the source of isolation, but not clinical phenotype.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Motility of Campylobacter concisus isolated from saliva, feces, and gut mucosal biopsies.APMIS. 2017 Mar;125(3):230-235. doi: 10.1111/apm.12655. Epub 2017 Jan 24. APMIS. 2017. PMID: 28116789
-
Comparative genomics of Campylobacter concisus: Analysis of clinical strains reveals genome diversity and pathogenic potential.Emerg Microbes Infect. 2018 Jun 26;7(1):116. doi: 10.1038/s41426-018-0118-x. Emerg Microbes Infect. 2018. PMID: 29946138 Free PMC article.
-
Delineation of genetic relatedness and population structure of oral and enteric Campylobacter concisus strains by analysis of housekeeping genes.Microbiology (Reading). 2015 Aug;161(8):1600-1612. doi: 10.1099/mic.0.000112. Epub 2015 May 22. Microbiology (Reading). 2015. PMID: 26002953
-
The Clinical Importance of Campylobacter concisus and Other Human Hosted Campylobacter Species.Front Cell Infect Microbiol. 2018 Jul 24;8:243. doi: 10.3389/fcimb.2018.00243. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30087857 Free PMC article. Review.
-
Campylobacter concisus and inflammatory bowel disease.World J Gastroenterol. 2014 Feb 7;20(5):1259-67. doi: 10.3748/wjg.v20.i5.1259. World J Gastroenterol. 2014. PMID: 24574800 Free PMC article. Review.
Cited by
-
Role of intestinal microbiota and metabolites in inflammatory bowel disease.Chin Med J (Engl). 2019 Jul 5;132(13):1610-1614. doi: 10.1097/CM9.0000000000000290. Chin Med J (Engl). 2019. PMID: 31090547 Free PMC article. Review.
-
Analysis of complete Campylobacter concisus genomes identifies genomospecies features, secretion systems and novel plasmids and their association with severe ulcerative colitis.Microb Genom. 2020 Nov;6(11):mgen000457. doi: 10.1099/mgen.0.000457. Microb Genom. 2020. PMID: 33111662 Free PMC article.
-
The role of the oral microbiome in obesity and metabolic disease: potential systemic implications and effects on taste perception.Nutr J. 2023 May 27;22(1):28. doi: 10.1186/s12937-023-00856-7. Nutr J. 2023. PMID: 37237407 Free PMC article. Review.
-
High genetic diversity in Campylobacter concisus isolates from patients with microscopic colitis.Gut Pathog. 2021 Jan 12;13(1):3. doi: 10.1186/s13099-020-00397-y. Gut Pathog. 2021. PMID: 33436056 Free PMC article.
-
Campylobacter concisus Impairs Sodium Absorption in Colonic Epithelium via ENaC Dysfunction and Claudin-8 Disruption.Int J Mol Sci. 2020 Jan 7;21(2):373. doi: 10.3390/ijms21020373. Int J Mol Sci. 2020. PMID: 31936044 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

