Targeting the SUMO pathway as a novel treatment for anaplastic thyroid cancer
- PMID: 29383121
- PMCID: PMC5777733
- DOI: 10.18632/oncotarget.21954
Targeting the SUMO pathway as a novel treatment for anaplastic thyroid cancer
Abstract
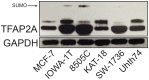
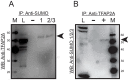
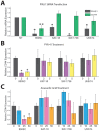
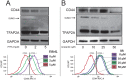
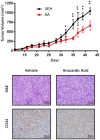
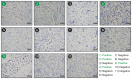
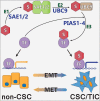
Cancer stem cells (CSCs) are expanded in anaplastic thyroid cancer (ATC) and standard treatment approaches have failed to improve survival, suggesting a need to specifically target the CSC population. Recent studies in breast and colorectal cancer demonstrated that inhibition of the SUMO pathway repressed CD44 and cleared the CSC population, mediated through SUMO-unconjugated TFAP2A. We sought to evaluate effects of inhibiting the SUMO pathway in ATC. ATC cell lines and primary ATC tumor samples were evaluated. The SUMO pathway was inhibited by knockdown of PIAS1 and use of SUMO inhibitors anacardic acid and PYR-41. The expression of TFAP2A in primary ATC was examined by immunohistochemistry. All ATC cell lines expressed TFAP2A but only 8505C expressed SUMO-conjugated TFAP2A. In 8505C only, inhibition of the SUMO pathway by knockdown of PIAS1 or treatment with SUMO inhibitors repressed expression of CD44 with a concomitant loss of SUMO-conjugated TFAP2A. The effect of SUMO inhibition on CD44 expression was dependent upon TFAP2A. Treatment with SUMO inhibitors resulted in a statistically improved tumor-free survival in mice harboring 8505C xenografts. An examination of primary ATC tissue determined that TFAP2A was expressed in 4 of 11 tumors surveyed. We conclude that inhibition of the SUMO pathway repressed the CSC population, delaying the outgrowth of tumor xenografts in ATC. The effect of SUMO inhibition was dependent upon expression of SUMO-conjugated TFAP2A, which may serve as a molecular marker for therapeutic effects of SUMO inhibitors. The findings provide pre-clinical evidence for development of SUMO inhibitors for the treatment of ATC.
Keywords: SUMO; TFAP2A; anaplastic thyroid cancer; sumoylation.
Conflict of interest statement
CONFLICTS OF INTEREST The authors have no conflicts of interest to disclose.
Figures





References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Charafe-Jauffret E, Ginestier C, Bertucci F, Cabaud O, Wicinski J, Finetti P, Josselin E, Adelaide J, Nguyen TT, Monville F, Jacquemier J, Thomassin-Piana J, Pinna G, et al. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res. 2013;73:7290–300. - PubMed
-
- Iqbal J, Chong PY, Tan PH. Breast cancer stem cells: an update. J Clin Pathol. 2013;66:485–90. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

