Structural basis of ribosomal peptide macrocyclization in plants
- PMID: 29384475
- PMCID: PMC5834244
- DOI: 10.7554/eLife.32955
Structural basis of ribosomal peptide macrocyclization in plants
Abstract
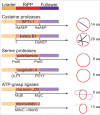
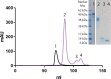
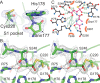
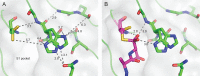
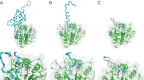
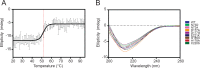
Constrained, cyclic peptides encoded by plant genes represent a new generation of drug leads. Evolution has repeatedly recruited the Cys-protease asparaginyl endopeptidase (AEP) to perform their head-to-tail ligation. These macrocyclization reactions use the substrates amino terminus instead of water to deacylate, so a peptide bond is formed. How solvent-exposed plant AEPs macrocyclize is poorly understood. Here we present the crystal structure of an active plant AEP from the common sunflower, Helianthus annuus. The active site contained electron density for a tetrahedral intermediate with partial occupancy that predicted a binding mode for peptide macrocyclization. By substituting catalytic residues we could alter the ratio of cyclic to acyclic products. Moreover, we showed AEPs from other species lacking cyclic peptides can perform macrocyclization under favorable pH conditions. This structural characterization of AEP presents a logical framework for engineering superior enzymes that generate macrocyclic peptide drug leads.
Keywords: Helianthus annuus; molecular biophysics; peptide macrocyclization; protease catalysis; structural biology; transpeptidation.
© 2018, Haywood et al.
Conflict of interest statement
JH, JS, AJ, SN, KS, ME, CB, JM No competing interests declared
Figures







Similar articles
-
Macrocyclization by asparaginyl endopeptidases.New Phytol. 2018 May;218(3):923-928. doi: 10.1111/nph.14511. Epub 2017 Mar 21. New Phytol. 2018. PMID: 28322452 Review.
-
Peptide macrocyclization by a bifunctional endoprotease.Chem Biol. 2015 May 21;22(5):571-82. doi: 10.1016/j.chembiol.2015.04.010. Epub 2015 May 7. Chem Biol. 2015. PMID: 25960260
-
The macrocyclizing protease butelase 1 remains autocatalytic and reveals the structural basis for ligase activity.Plant J. 2019 Jun;98(6):988-999. doi: 10.1111/tpj.14293. Epub 2019 Mar 28. Plant J. 2019. PMID: 30790358
-
Segmental isotopic labeling of a single-domain globular protein without any refolding step by an asparaginyl endopeptidase.FEBS Lett. 2017 May;591(9):1285-1294. doi: 10.1002/1873-3468.12640. Epub 2017 Apr 20. FEBS Lett. 2017. PMID: 28369872
-
Plant asparaginyl endopeptidases and their structural determinants of function.Biochem Soc Trans. 2021 Apr 30;49(2):965-976. doi: 10.1042/BST20200908. Biochem Soc Trans. 2021. PMID: 33666219 Free PMC article. Review.
Cited by
-
A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.Nat Commun. 2022 Sep 22;13(1):5563. doi: 10.1038/s41467-022-33185-0. Nat Commun. 2022. PMID: 36137996 Free PMC article.
-
Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.Plant Cell. 2022 Nov 29;34(12):4936-4949. doi: 10.1093/plcell/koac281. Plant Cell. 2022. PMID: 36099055 Free PMC article.
-
A suite of kinetically superior AEP ligases can cyclise an intrinsically disordered protein.Sci Rep. 2019 Jul 25;9(1):10820. doi: 10.1038/s41598-019-47273-7. Sci Rep. 2019. PMID: 31346249 Free PMC article.
-
Structural determinants for peptide-bond formation by asparaginyl ligases.Proc Natl Acad Sci U S A. 2019 Jun 11;116(24):11737-11746. doi: 10.1073/pnas.1818568116. Epub 2019 May 23. Proc Natl Acad Sci U S A. 2019. PMID: 31123145 Free PMC article.
-
Asparaginyl endopeptidases: enzymology, applications and limitations.Org Biomol Chem. 2021 Jun 16;19(23):5048-5062. doi: 10.1039/d1ob00608h. Org Biomol Chem. 2021. PMID: 34037066 Free PMC article. Review.
References
-
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/C2NP20085F. - DOI - PMC - PubMed
-
- Bergmann M, Fruton JS. Some synthetic and hydrolytic experiments with chymotrypsin. The Journal of Biological Chemistry. 1938;124:321–329.
-
- Bhardwaj G, Mulligan VK, Bahl CD, Gilmore JM, Harvey PJ, Cheneval O, Buchko GW, Pulavarti SV, Kaas Q, Eletsky A, Huang PS, Johnsen WA, Greisen PJ, Rocklin GJ, Song Y, Linsky TW, Watkins A, Rettie SA, Xu X, Carter LP, Bonneau R, Olson JM, Coutsias E, Correnti CE, Szyperski T, Craik DJ, Baker D. Accurate de novo design of hyperstable constrained peptides. Nature. 2016;538:329–335. doi: 10.1038/nature19791. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

