The intra- and extraluminal appendiceal microbiome in pediatric patients: A comparative study
- PMID: 29384958
- PMCID: PMC6393148
- DOI: 10.1097/MD.0000000000009518
The intra- and extraluminal appendiceal microbiome in pediatric patients: A comparative study
Abstract
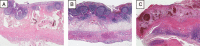
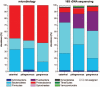
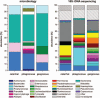
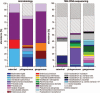
Intestinal microbiota is involved in metabolic processes and the pathophysiology of various gastrointestinal disorders. We aimed to characterize the microbiome of the appendix in acute pediatric appendicitis comparing extraluminal and intraluminal samples.Between January and June 2015, 29 children (3-17 years, mean age 10.7 ± 3.4 years, sex M:F = 2.6:1) undergoing laparoscopic appendectomy for acute appendicitis were prospectively included in the study. Samples for bacterial cultures (n = 29) and 16S ribosomal desoxyribonucleic acid (rDNA) sequencing (randomly chosen n = 16/29) were taken intracorporeally from the appendiceal surface before preparation ("extraluminal") and from the appendiceal lumen after removal ("intraluminal"). The degree of inflammation was histologically classified into catarrhal, phlegmonous, and gangrenous appendicitis.Seventeen bacterial species were cultivated in 28 of 29 intraluminal samples and 4 species were cultivated in 2 of 29 extraluminal samples. Using 16S rDNA sequencing, 267 species were detected in intraluminal but none in extraluminal samples. Abundance and diversity of detected species differed significantly between histological groups of acute appendicitis in bacterial cultures (P = .001), but not after 16S rDNA sequencing.The appendiceal microbiome showed a high diversity in acute pediatric appendicitis. The intraluminal microbial composition differed significantly depending on the degree of inflammation. As bacteria were rarely found extraluminally by culture and not at all by sequencing, the inflammation in acute appendicitis may start inside the appendix and spread transmurally.
Copyright © 2017 The Authors. Published by Wolters Kluwer Health, Inc. All rights reserved.
Figures
Similar articles
-
The oral microbiome-the relevant reservoir for acute pediatric appendicitis?Int J Colorectal Dis. 2018 Feb;33(2):209-218. doi: 10.1007/s00384-017-2948-8. Epub 2017 Dec 22. Int J Colorectal Dis. 2018. PMID: 29273882
-
Acute appendicitis in children is associated with an abundance of bacteria from the phylum Fusobacteria.J Pediatr Surg. 2014 Mar;49(3):441-6. doi: 10.1016/j.jpedsurg.2013.06.026. J Pediatr Surg. 2014. PMID: 24650474
-
Microbiota of Children With Complex Appendicitis: Different Composition and Diversity of The Microbiota in Children With Complex Compared With Simple Appendicitis.Pediatr Infect Dis J. 2019 Oct;38(10):1054-1060. doi: 10.1097/INF.0000000000002434. Pediatr Infect Dis J. 2019. PMID: 31568143
-
Review of the histopathological findings in appendices removed for acute appendicitis in Nigerians.J R Coll Surg Edinb. 1991 Aug;36(4):245-8. J R Coll Surg Edinb. 1991. PMID: 1941741 Review.
-
Clinical manifestations of appendiceal pinworms in children: an institutional experience and a review of the literature.Pediatr Surg Int. 2004 May;20(5):372-5. doi: 10.1007/s00383-004-1151-5. Epub 2004 May 13. Pediatr Surg Int. 2004. PMID: 15141320 Review.
Cited by
-
Microbiota Assessment of Pediatric Simple and Complex Acute Appendicitis.Medicina (Kaunas). 2022 Aug 23;58(9):1144. doi: 10.3390/medicina58091144. Medicina (Kaunas). 2022. PMID: 36143821 Free PMC article.
-
Comparison of the efficacy of carbapenems and cephalosporins for postoperative treatment of perforated appendicitis in children.World J Gastrointest Surg. 2025 Apr 27;17(4):104712. doi: 10.4240/wjgs.v17.i4.104712. World J Gastrointest Surg. 2025. PMID: 40291900 Free PMC article.
-
Acute appendicitis is associated with appendiceal microbiome changes including elevated Campylobacter jejuni levels.BMJ Open Gastroenterol. 2020 Jun;7(1):e000412. doi: 10.1136/bmjgast-2020-000412. BMJ Open Gastroenterol. 2020. PMID: 32499276 Free PMC article.
-
The microbiome in adult acute appendicitis.Gut Microbiome (Camb). 2022 Aug 4;3:e8. doi: 10.1017/gmb.2022.7. eCollection 2022. Gut Microbiome (Camb). 2022. PMID: 39295777 Free PMC article.
-
Appendiceal microbiome in uncomplicated and complicated acute appendicitis: A prospective cohort study.PLoS One. 2022 Oct 14;17(10):e0276007. doi: 10.1371/journal.pone.0276007. eCollection 2022. PLoS One. 2022. PMID: 36240181 Free PMC article.
References
-
- Birnbaum BA, Wilson SR. Appendicitis at the millennium. Radiology 2000;215:337–48. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

