Disseminating Metaproteomic Informatics Capabilities and Knowledge Using the Galaxy-P Framework
- PMID: 29385081
- PMCID: PMC5874766
- DOI: 10.3390/proteomes6010007
Disseminating Metaproteomic Informatics Capabilities and Knowledge Using the Galaxy-P Framework
Abstract
The impact of microbial communities, also known as the microbiome, on human health and the environment is receiving increased attention. Studying translated gene products (proteins) and comparing metaproteomic profiles may elucidate how microbiomes respond to specific environmental stimuli, and interact with host organisms. Characterizing proteins expressed by a complex microbiome and interpreting their functional signature requires sophisticated informatics tools and workflows tailored to metaproteomics. Additionally, there is a need to disseminate these informatics resources to researchers undertaking metaproteomic studies, who could use them to make new and important discoveries in microbiome research. The Galaxy for proteomics platform (Galaxy-P) offers an open source, web-based bioinformatics platform for disseminating metaproteomics software and workflows. Within this platform, we have developed easily-accessible and documented metaproteomic software tools and workflows aimed at training researchers in their operation and disseminating the tools for more widespread use. The modular workflows encompass the core requirements of metaproteomic informatics: (a) database generation; (b) peptide spectral matching; (c) taxonomic analysis and (d) functional analysis. Much of the software available via the Galaxy-P platform was selected, packaged and deployed through an online metaproteomics "Contribution Fest" undertaken by a unique consortium of expert software developers and users from the metaproteomics research community, who have co-authored this manuscript. These resources are documented on GitHub and freely available through the Galaxy Toolshed, as well as a publicly accessible metaproteomics gateway Galaxy instance. These documented workflows are well suited for the training of novice metaproteomics researchers, through online resources such as the Galaxy Training Network, as well as hands-on training workshops. Here, we describe the metaproteomics tools available within these Galaxy-based resources, as well as the process by which they were selected and implemented in our community-based work. We hope this description will increase access to and utilization of metaproteomics tools, as well as offer a framework for continued community-based development and dissemination of cutting edge metaproteomics software.
Keywords: Galaxy platform; bioinformatics; community development; functional microbiome; mass spectrometry; metaproteomics; software workflow development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
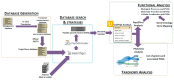





References
-
- Maier T.V., Lucio M., Lee L.H., VerBerkmoes N.C., Brislawn C.J., Bernhardt J., Lamendella R., McDermott J.E., Bergeron N., Heinzmann S.S., et al. Impact of Dietary Resistant Starch on the Human Gut Microbiome, Metaproteome, and Metabolome. mBio. 2017;8:1343–1417. doi: 10.1128/mBio.01343-17. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

