Translational Selection for Speed Is Not Sufficient to Explain Variation in Bacterial Codon Usage Bias
- PMID: 29385509
- PMCID: PMC5800062
- DOI: 10.1093/gbe/evy018
Translational Selection for Speed Is Not Sufficient to Explain Variation in Bacterial Codon Usage Bias
Abstract
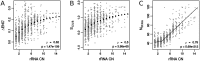
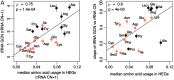
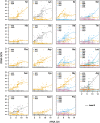
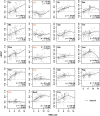
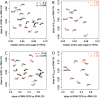
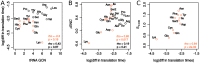
Increasing growth rate across bacteria strengthens selection for faster translation, concomitantly increasing the total number of tRNA genes and codon usage bias (CUB: enrichment of specific synonymous codons in highly expressed genes). Typically, enriched codons are translated by tRNAs with higher gene copy numbers (GCN). A model of tRNA-CUB coevolution based on fast growth-associated selection on translational speed recapitulates these patterns. A key untested implication of the coevolution model is that translational selection should favor higher tRNA GCN for more frequently used amino acids, potentially weakening the effect of growth-associated selection on CUB. Surprisingly, we find that CUB saturates with increasing growth rate across γ-proteobacteria, even as the number of tRNA genes continues to increase. As predicted, amino acid-specific tRNA GCN is positively correlated with the usage of corresponding amino acids, but there is no correlation between growth rate associated changes in CUB and amino acid usage. Instead, we find that some amino acids-cysteine and those in the NNA/G codon family-show weak CUB that does not increase with growth rate, despite large variation in the corresponding tRNA GCN. We suggest that amino acid-specific variation in CUB is not explained by tRNA GCN because GCN does not influence the difference between translation times of synonymous codons as expected. Thus, selection on translational speed alone cannot fully explain quantitative variation in overall or amino acid-specific CUB, suggesting a significant role for other functional constraints and amino acid-specific codon features.
Keywords: amino acid usage; effective number of codons; growth rate; rRNA copy number; tRNA gene copy number.
© The Author(s) 2018. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage.Mol Biol Evol. 2008 Nov;25(11):2279-91. doi: 10.1093/molbev/msn173. Epub 2008 Aug 6. Mol Biol Evol. 2008. PMID: 18687657
-
Coevolution of codon usage and transfer RNA abundance.Nature. 1987 Feb 19-25;325(6106):728-30. doi: 10.1038/325728a0. Nature. 1987. PMID: 2434856
-
Gene expression levels are correlated with synonymous codon usage, amino acid composition, and gene architecture in the red flour beetle, Tribolium castaneum.Mol Biol Evol. 2012 Dec;29(12):3755-66. doi: 10.1093/molbev/mss184. Epub 2012 Jul 23. Mol Biol Evol. 2012. PMID: 22826459 Free PMC article.
-
Evolutionary tuning impacts the design of bacterial tRNAs for the incorporation of unnatural amino acids by ribosomes.Curr Opin Chem Biol. 2018 Oct;46:138-145. doi: 10.1016/j.cbpa.2018.07.016. Epub 2018 Jul 27. Curr Opin Chem Biol. 2018. PMID: 30059836 Free PMC article. Review.
-
The Yin and Yang of codon usage.Hum Mol Genet. 2016 Oct 1;25(R2):R77-R85. doi: 10.1093/hmg/ddw207. Epub 2016 Jun 27. Hum Mol Genet. 2016. PMID: 27354349 Free PMC article. Review.
Cited by
-
Synonymous codon substitutions modulate transcription and translation of a divergent upstream gene by modulating antisense RNA production.Proc Natl Acad Sci U S A. 2024 Sep 3;121(36):e2405510121. doi: 10.1073/pnas.2405510121. Epub 2024 Aug 27. Proc Natl Acad Sci U S A. 2024. PMID: 39190361 Free PMC article.
-
Analyzing the Evolution and Host Adaptation of the Rabies Virus from the Perspective of Codon Usage Bias.Transbound Emerg Dis. 2023 Oct 10;2023:4667253. doi: 10.1155/2023/4667253. eCollection 2023. Transbound Emerg Dis. 2023. PMID: 40303686 Free PMC article.
-
The layered costs and benefits of translational redundancy.Elife. 2023 Mar 2;12:e81005. doi: 10.7554/eLife.81005. Elife. 2023. PMID: 36862572 Free PMC article.
-
Can Protein Expression Be Regulated by Modulation of tRNA Modification Profiles?Biochemistry. 2019 Feb 5;58(5):355-362. doi: 10.1021/acs.biochem.8b01035. Epub 2018 Dec 18. Biochemistry. 2019. PMID: 30511849 Free PMC article.
-
The Key Parameters that Govern Translation Efficiency.Cell Syst. 2020 Feb 26;10(2):183-192.e6. doi: 10.1016/j.cels.2019.12.003. Epub 2020 Jan 15. Cell Syst. 2020. PMID: 31954660 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

