TelAP1 links telomere complexes with developmental expression site silencing in African trypanosomes
- PMID: 29385523
- PMCID: PMC5888660
- DOI: 10.1093/nar/gky028
TelAP1 links telomere complexes with developmental expression site silencing in African trypanosomes
Abstract
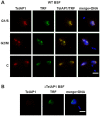
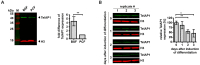
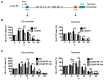
During its life cycle, Trypanosoma brucei shuttles between a mammalian host and the tsetse fly vector. In the mammalian host, immune evasion of T. brucei bloodstream form (BSF) cells relies on antigenic variation, which includes monoallelic expression and periodic switching of variant surface glycoprotein (VSG) genes. The active VSG is transcribed from only 1 of the 15 subtelomeric expression sites (ESs). During differentiation from BSF to the insect-resident procyclic form (PCF), the active ES is transcriptionally silenced. We used mass spectrometry-based interactomics to determine the composition of telomere protein complexes in T. brucei BSF and PCF stages to learn more about the structure and functions of telomeres in trypanosomes. Our data suggest a different telomere complex composition in the two forms of the parasite. One of the novel telomere-associated proteins, TelAP1, forms a complex with telomeric proteins TbTRF, TbRAP1 and TbTIF2 and influences ES silencing kinetics during developmental differentiation.
Figures





Similar articles
-
TelAP2 links TelAP1 to the telomere complex in Trypanosoma brucei.Sci Rep. 2024 Dec 16;14(1):30493. doi: 10.1038/s41598-024-81972-0. Sci Rep. 2024. PMID: 39681615 Free PMC article.
-
Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids.Nucleic Acids Res. 2017 Jun 2;45(10):5785-5796. doi: 10.1093/nar/gkx184. Nucleic Acids Res. 2017. PMID: 28334836 Free PMC article.
-
Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes.Proc Natl Acad Sci U S A. 2015 May 26;112(21):E2803-12. doi: 10.1073/pnas.1501206112. Epub 2015 May 11. Proc Natl Acad Sci U S A. 2015. PMID: 25964327 Free PMC article.
-
The VSG expression sites of Trypanosoma brucei: multipurpose tools for the adaptation of the parasite to mammalian hosts.Mol Biochem Parasitol. 2001 Apr 25;114(1):1-16. doi: 10.1016/s0166-6851(01)00242-0. Mol Biochem Parasitol. 2001. PMID: 11356509 Review.
-
A Host-Pathogen Interaction Reduced to First Principles: Antigenic Variation in T. brucei.Results Probl Cell Differ. 2015;57:23-46. doi: 10.1007/978-3-319-20819-0_2. Results Probl Cell Differ. 2015. PMID: 26537376 Review.
Cited by
-
Unwrap RAP1's Mystery at Kinetoplastid Telomeres.Biomolecules. 2024 Jan 4;14(1):67. doi: 10.3390/biom14010067. Biomolecules. 2024. PMID: 38254667 Free PMC article. Review.
-
Escaping the immune system by DNA repair and recombination in African trypanosomes.Open Biol. 2019 Nov 29;9(11):190182. doi: 10.1098/rsob.190182. Epub 2019 Nov 13. Open Biol. 2019. PMID: 31718509 Free PMC article. Review.
-
DNA double strand break position leads to distinct gene expression changes and regulates VSG switching pathway choice.PLoS Pathog. 2021 Nov 12;17(11):e1010038. doi: 10.1371/journal.ppat.1010038. eCollection 2021 Nov. PLoS Pathog. 2021. PMID: 34767618 Free PMC article.
-
A novel SNF2 ATPase complex in Trypanosoma brucei with a role in H2A.Z-mediated chromatin remodelling.PLoS Pathog. 2022 Jun 8;18(6):e1010514. doi: 10.1371/journal.ppat.1010514. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35675371 Free PMC article.
-
The developmental hierarchy and scarcity of replicative slender trypanosomes in blood challenges their role in infection maintenance.Proc Natl Acad Sci U S A. 2023 Oct 17;120(42):e2306848120. doi: 10.1073/pnas.2306848120. Epub 2023 Oct 12. Proc Natl Acad Sci U S A. 2023. PMID: 37824530 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

