Differential Glycosylation and Modulation of Camel and Human HSP Isoforms in Response to Thermal and Hypoxic Stresses
- PMID: 29385708
- PMCID: PMC5855624
- DOI: 10.3390/ijms19020402
Differential Glycosylation and Modulation of Camel and Human HSP Isoforms in Response to Thermal and Hypoxic Stresses
Abstract
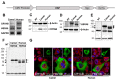
Increased expression of heat shock proteins (HSPs) following heat stress or other stress conditions is a common physiological response in almost all living organisms. Modification of cytosolic proteins including HSPs by O-GlcNAc has been shown to enhance their capabilities for counteracting lethal levels of cellular stress. Since HSPs are key players in stress resistance and protein homeostasis, we aimed to analyze their forms at the cellular and molecular level using camel and human HSPs as models for efficient and moderate thermotolerant mammals, respectively. In this study, we cloned the cDNA encoding two inducible HSP members, HSPA6 and CRYAB from both camel (Camelus dromedarius) and human in a Myc-tagged mammalian expression vector. Expression of these chaperones in COS-1 cells revealed protein bands of approximately 25-kDa for both camel and human CRYAB and 70-kDa for camel HSPA6 and its human homologue. While localization and trafficking of the camel and human HSPs revealed similar cytosolic localization, we could demonstrate altered glycan structure between camel and human HSPA6. Interestingly, the glycoform of camel HSPA6 was rapidly formed and stabilized under normal and stress culture conditions whereas human HSPA6 reacted differently under similar thermal and hypoxic stress conditions. Our data suggest that efficient glycosylation of camel HSPA6 is among the mechanisms that provide camelids with a superior capability for alleviating stressful environmental circumstances.
Keywords: CRYAB; HSPA6; O-GlcNAc; camel; heat shock proteins; heat stress; hypoxia; protein expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Cellular and Molecular Adaptation of Arabian Camel to Heat Stress.Front Genet. 2019 Jun 19;10:588. doi: 10.3389/fgene.2019.00588. eCollection 2019. Front Genet. 2019. PMID: 31275361 Free PMC article. Review.
-
Molecular cloning and characterization of cDNA encoding a putative stress-induced heat-shock protein from Camelus dromedarius.Int J Mol Sci. 2011;12(7):4214-36. doi: 10.3390/ijms12074214. Epub 2011 Jun 27. Int J Mol Sci. 2011. PMID: 21845074 Free PMC article.
-
Localization of heat shock protein HSPA6 (HSP70B') to sites of transcription in cultured differentiated human neuronal cells following thermal stress.J Neurochem. 2014 Dec;131(6):743-54. doi: 10.1111/jnc.12970. Epub 2014 Dec 2. J Neurochem. 2014. PMID: 25319762
-
Functional organization of hsp70 cluster in camel (Camelus dromedarius) and other mammals.PLoS One. 2011;6(11):e27205. doi: 10.1371/journal.pone.0027205. Epub 2011 Nov 9. PLoS One. 2011. PMID: 22096537 Free PMC article.
-
Changes in skeletal muscle heat shock proteins: pathological significance.Front Biosci. 2001 Jan 1;6:D12-25. doi: 10.2741/liu. Front Biosci. 2001. PMID: 11145923 Review.
Cited by
-
O-GlcNAcylation: the "stress and nutrition receptor" in cell stress response.Cell Stress Chaperones. 2021 Mar;26(2):297-309. doi: 10.1007/s12192-020-01177-y. Epub 2020 Nov 7. Cell Stress Chaperones. 2021. PMID: 33159661 Free PMC article. Review.
-
O-GlcNAcylation: a pro-survival response to acute stress in the cardiovascular and central nervous systems.Eur J Med Res. 2024 Mar 16;29(1):174. doi: 10.1186/s40001-024-01773-z. Eur J Med Res. 2024. PMID: 38491477 Free PMC article. Review.
-
Cellular and Molecular Adaptation of Arabian Camel to Heat Stress.Front Genet. 2019 Jun 19;10:588. doi: 10.3389/fgene.2019.00588. eCollection 2019. Front Genet. 2019. PMID: 31275361 Free PMC article. Review.
-
Lectin binding to pectoral fin of neonate little skates reared under ambient and projected-end-of-century temperature regimes.J Morphol. 2024 May;285(5):e21698. doi: 10.1002/jmor.21698. J Morphol. 2024. PMID: 38669130 Free PMC article.
-
The molecular chaperone Hsp70 from the thermotolerant Diptera species differs from the Drosophila paralog in its thermostability and higher refolding capacity at extreme temperatures.Cell Stress Chaperones. 2019 Nov;24(6):1163-1173. doi: 10.1007/s12192-019-01038-3. Epub 2019 Oct 30. Cell Stress Chaperones. 2019. PMID: 31664698 Free PMC article.
References
-
- Schmidt-Nilsen K. Animals of the Deserts. Nauka; Moscow, Russia: 1972.
-
- Ouajd S., Kamel B. Physiological particularities of dromedary (Camelus dromedarius) and experimental implications. Scand. J. Lab. Anim. Sci. 2009;36:19–29.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

