The Diverging Routes of BORIS and CTCF: An Interactomic and Phylogenomic Analysis
- PMID: 29385718
- PMCID: PMC5871936
- DOI: 10.3390/life8010004
The Diverging Routes of BORIS and CTCF: An Interactomic and Phylogenomic Analysis
Abstract
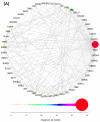
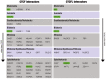
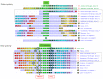
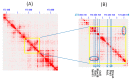
The CCCTC-binding factor (CTCF) is multi-functional, ubiquitously expressed, and highly conserved from Drosophila to human. It has important roles in transcriptional insulation and the formation of a high-dimensional chromatin structure. CTCF has a paralog called "Brother of Regulator of Imprinted Sites" (BORIS) or "CTCF-like" (CTCFL). It binds DNA at sites similar to those of CTCF. However, the expression profiles of the two proteins are quite different. We investigated the evolutionary trajectories of the two proteins after the duplication event using a phylogenomic and interactomic approach. We find that CTCF has 52 direct interaction partners while CTCFL only has 19. Almost all interactors already existed before the emergence of CTCF and CTCFL. The unique secondary loss of CTCF from several nematodes is paralleled by a loss of two of its interactors, the polycomb repressive complex subunit SuZ12 and the multifunctional transcription factor TYY1. In contrast to earlier studies reporting the absence of BORIS from birds, we present evidence for a multigene synteny block containing CTCFL that is conserved in mammals, reptiles, and several species of birds, indicating that not the entire lineage of birds experienced a loss of CTCFL. Within this synteny block, BORIS and its genomic neighbors seem to be partitioned into two nested chromatin loops. The high expression of SPO11, RAE1, RBM38, and PMEPA1 in male tissues suggests a possible link between CTCFL, meiotic recombination, and fertility-associated phenotypes. Using the 65,700 exomes and the 1000 genomes data, we observed a higher number of intergenic, non-synonymous, and loss-of-function mutations in CTCFL than in CTCF, suggesting a reduced strength of purifying selection, perhaps due to less functional constraint.
Keywords: Amniotes; Bilateria; CTCF; chromatin loops; gene duplication; natural selection; polymorphism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Hypomethylation of the CTCFL/BORIS promoter and aberrant expression during endometrial cancer progression suggests a role as an Epi-driver gene.Oncotarget. 2014 Feb 28;5(4):1052-61. doi: 10.18632/oncotarget.1697. Oncotarget. 2014. PMID: 24658009 Free PMC article.
-
The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes.PLoS Genet. 2008 Aug 29;4(8):e1000169. doi: 10.1371/journal.pgen.1000169. PLoS Genet. 2008. PMID: 18769711 Free PMC article.
-
Discovering a binary CTCF code with a little help from BORIS.Nucleus. 2018 Jan 1;9(1):33-41. doi: 10.1080/19491034.2017.1394536. Epub 2017 Dec 5. Nucleus. 2018. PMID: 29077515 Free PMC article. Review.
-
Early vertebrate origin of CTCFL, a CTCF paralog, revealed by proximity-guided shark genome scaffolding.Sci Rep. 2020 Sep 3;10(1):14629. doi: 10.1038/s41598-020-71602-w. Sci Rep. 2020. PMID: 32884037 Free PMC article.
-
Interaction of CTCF and CTCFL in genome regulation through chromatin architecture during the spermatogenesis and carcinogenesis.PeerJ. 2024 Oct 15;12:e18240. doi: 10.7717/peerj.18240. eCollection 2024. PeerJ. 2024. PMID: 39430552 Free PMC article. Review.
Cited by
-
A common genomic code for chromatin architecture and recombination landscape.PLoS One. 2019 Mar 13;14(3):e0213278. doi: 10.1371/journal.pone.0213278. eCollection 2019. PLoS One. 2019. PMID: 30865674 Free PMC article.
-
Interactome mapping defines BRG1, a component of the SWI/SNF chromatin remodeling complex, as a new partner of the transcriptional regulator CTCF.J Biol Chem. 2019 Jan 18;294(3):861-873. doi: 10.1074/jbc.RA118.004882. Epub 2018 Nov 20. J Biol Chem. 2019. PMID: 30459231 Free PMC article.
-
Molecular Lesions of Insulator CTCF and Its Paralogue CTCFL (BORIS) in Cancer: An Analysis from Published Genomic Studies.High Throughput. 2018 Oct 1;7(4):30. doi: 10.3390/ht7040030. High Throughput. 2018. PMID: 30275357 Free PMC article.
-
Zmym4 is required for early cranial gene expression and craniofacial cartilage formation.Front Cell Dev Biol. 2023 Oct 3;11:1274788. doi: 10.3389/fcell.2023.1274788. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37854072 Free PMC article.
References
-
- Filippova G.N., Qi C.F., Ulmer J.E., Moore J.M., Ward M.D., Hu Y.J., Loukinov D.I., Pugacheva E.M., Klenova E.M., Grundy P.E., et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res. 2002;62:48–52. - PubMed
-
- Lobanenkov V.V., Nicolas R.H., Adler V.V., Paterson H., Klenova E.M., Polotskaja A.V., Goodwin G.H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC motif in the 5′-flanking sequence of the chicken c-Myc gene. Oncogene. 1990;5:1743–1753. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

