Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases
- PMID: 29385775
- PMCID: PMC5855634
- DOI: 10.3390/ijms19020412
Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases
Abstract
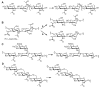
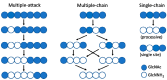
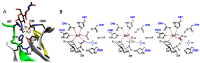
Carbohydrate esterases family 4 (CE4 enzymes) includes chitin and peptidoglycan deacetylases, acetylxylan esterases, and poly-N-acetylglucosamine deacetylases that act on structural polysaccharides, altering their physicochemical properties, and participating in diverse biological functions. Chitin and peptidoglycan deacetylases are not only involved in cell wall morphogenesis and remodeling in fungi and bacteria, but they are also used by pathogenic microorganisms to evade host defense mechanisms. Likewise, biofilm formation in bacteria requires partial deacetylation of extracellular polysaccharides mediated by poly-N-acetylglucosamine deacetylases. Such biological functions make these enzymes attractive targets for drug design against pathogenic fungi and bacteria. On the other side, acetylxylan esterases deacetylate plant cell wall complex xylans to make them accessible to hydrolases, making them attractive biocatalysts for biomass utilization. CE4 family members are metal-dependent hydrolases. They are highly specific for their particular substrates, and show diverse modes of action, exhibiting either processive, multiple attack, or patterned deacetylation mechanisms. However, the determinants of substrate specificity remain poorly understood. Here, we review the current knowledge on the structure, activity, and specificity of CE4 enzymes, focusing on chitin deacetylases and related enzymes active on N-acetylglucosamine-containing oligo and polysaccharides.
Keywords: carbohydrate esterases; chitin deacetylases; chitooligosaccharides; chitosan; deacetylation pattern; peptidoglycan; structure; substrate specificity.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study, in the writing of the manuscript, and in the decision to publish the results.
Figures



Similar articles
-
Structure-function relationships underlying the dual N-acetylmuramic and N-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.J Biol Chem. 2019 Dec 13;294(50):19066-19080. doi: 10.1074/jbc.RA119.009510. Epub 2019 Nov 5. J Biol Chem. 2019. PMID: 31690626 Free PMC article.
-
The molecular structure, biological roles, and inhibition of plant pathogenic fungal chitin deacetylases.Front Plant Sci. 2024 Jan 9;14:1335646. doi: 10.3389/fpls.2023.1335646. eCollection 2023. Front Plant Sci. 2024. PMID: 38264029 Free PMC article. Review.
-
Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases.J Biol Chem. 2006 Apr 21;281(16):10968-75. doi: 10.1074/jbc.M513066200. Epub 2006 Jan 23. J Biol Chem. 2006. PMID: 16431911
-
Assessment of the properties of chitin deacetylases showing different enzymatic action patterns.J Mol Graph Model. 2019 May;88:41-48. doi: 10.1016/j.jmgm.2019.01.002. Epub 2019 Jan 9. J Mol Graph Model. 2019. PMID: 30660982
-
Chitin deacetylases: properties and applications.Mar Drugs. 2010 Jan 14;8(1):24-46. doi: 10.3390/md8010024. Mar Drugs. 2010. PMID: 20161969 Free PMC article. Review.
Cited by
-
Characterization of the Specific Mode of Action of a Chitin Deacetylase and Separation of the Partially Acetylated Chitosan Oligosaccharides.Mar Drugs. 2019 Jan 22;17(2):74. doi: 10.3390/md17020074. Mar Drugs. 2019. PMID: 30678277 Free PMC article.
-
Genome and Secretome Analysis of Staphylotrichum longicolleum DSM105789 Cultured on Agro-Residual and Chitinous Biomass.Microorganisms. 2021 Jul 25;9(8):1581. doi: 10.3390/microorganisms9081581. Microorganisms. 2021. PMID: 34442660 Free PMC article.
-
Variations of the NodB Architecture Are Attuned to Functional Specificities into and beyond the Carbohydrate Esterase Family 4.Biomolecules. 2024 Mar 8;14(3):325. doi: 10.3390/biom14030325. Biomolecules. 2024. PMID: 38540745 Free PMC article.
-
A new strain of Rhodococcus indonesiensis T22.7.1T and its functional potential for deacetylation of chitin and chitooligsaccharides.Front Microbiol. 2024 Jul 24;15:1427143. doi: 10.3389/fmicb.2024.1427143. eCollection 2024. Front Microbiol. 2024. PMID: 39113839 Free PMC article.
-
Functional analysis of the N-terminal region of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.FEBS Open Bio. 2022 Oct;12(10):1875-1885. doi: 10.1002/2211-5463.13476. Epub 2022 Sep 20. FEBS Open Bio. 2022. PMID: 36054591 Free PMC article.
References
-
- Nakamura A.M., Nascimento A.S., Polikarpov I. Structural diversity of carbohydrate esterases. Biotechnol. Res. Innov. 2017;1:35–51. doi: 10.1016/j.biori.2017.02.001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

