The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization
- PMID: 29386355
- PMCID: PMC5857997
- DOI: 10.1074/jbc.RA117.001294
The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization
Abstract
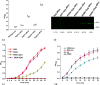
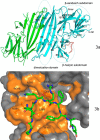
PRKR-like endoplasmic reticulum kinase (PERK) is one of the major sensor proteins that detect protein folding imbalances during endoplasmic reticulum (ER) stress. However, it remains unclear how ER stress activates PERK to initiate a downstream unfolded protein response (UPR). Here, we found that PERK's luminal domain can recognize and selectively interact with misfolded proteins but not with native proteins. Screening a phage-display library, we identified a peptide substrate, P16, of the PERK luminal domain and confirmed that P16 efficiently competes with misfolded proteins for binding this domain. To unravel the mechanism by which the PERK luminal domain interacts with misfolded proteins, we determined the crystal structure of the bovine PERK luminal domain complexed with P16 to 2.8-Å resolution. The structure revealed that PERK's luminal domain binds the peptide through a conserved hydrophobic groove. Substitutions within hydrophobic regions of the PERK luminal domain abolished the binding between PERK and misfolded proteins. We also noted that peptide binding results in major conformational changes in the PERK luminal domain that may favor PERK oligomerization. The structure of the PERK luminal domain-P16 complex suggested stacking of the luminal domain that leads to PERK oligomerization and activation via autophosphorylation after ligand binding. Collectively, our structural and biochemical results strongly support a ligand-driven model in which the PERK luminal domain interacts directly with misfolded proteins to induce PERK oligomerization and activation, resulting in ER stress signaling and the UPR.
Keywords: ER stress activation; PERK; crystal structure; crystallography; eukaryotic translation initiation factor 2α kinase 3; signaling; stress response; structural biology; unfolded protein response (UPR).
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

