Human migration and the spread of malaria parasites to the New World
- PMID: 29386521
- PMCID: PMC5792595
- DOI: 10.1038/s41598-018-19554-0
Human migration and the spread of malaria parasites to the New World
Abstract
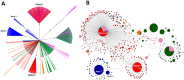
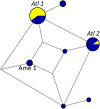
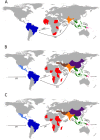
We examined the mitogenomes of a large global collection of human malaria parasites to explore how and when Plasmodium falciparum and P. vivax entered the Americas. We found evidence of a significant contribution of African and South Asian lineages to present-day New World malaria parasites with additional P. vivax lineages appearing to originate from Melanesia that were putatively carried by the Australasian peoples who contributed genes to Native Americans. Importantly, mitochondrial lineages of the P. vivax-like species P. simium are shared by platyrrhine monkeys and humans in the Atlantic Forest ecosystem, but not across the Amazon, which most likely resulted from one or a few recent human-to-monkey transfers. While enslaved Africans were likely the main carriers of P. falciparum mitochondrial lineages into the Americas after the conquest, additional parasites carried by Australasian peoples in pre-Columbian times may have contributed to the extensive diversity of extant local populations of P. vivax.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Why Plasmodium vivax and Plasmodium falciparum are so different? A tale of two clades and their species diversities.Malar J. 2022 May 3;21(1):139. doi: 10.1186/s12936-022-04130-9. Malar J. 2022. PMID: 35505356 Free PMC article. Review.
-
Genetic Markers of Adaptation of Plasmodium falciparum to Transmission by American Vectors Identified in the Genomes of Parasites from Haiti and South America.mSphere. 2020 Oct 21;5(5):e00937-20. doi: 10.1128/mSphere.00937-20. mSphere. 2020. PMID: 33087522 Free PMC article.
-
Ancient Plasmodium genomes shed light on the history of human malaria.Nature. 2024 Jul;631(8019):125-133. doi: 10.1038/s41586-024-07546-2. Epub 2024 Jun 12. Nature. 2024. PMID: 38867050 Free PMC article.
-
Origins of human malaria: rare genomic changes and full mitochondrial genomes confirm the relationship of Plasmodium falciparum to other mammalian parasites but complicate the origins of Plasmodium vivax.Mol Biol Evol. 2008 Jun;25(6):1192-8. doi: 10.1093/molbev/msn069. Epub 2008 Mar 21. Mol Biol Evol. 2008. PMID: 18359945 Free PMC article.
-
Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax.Int J Parasitol. 2017 Feb;47(2-3):87-97. doi: 10.1016/j.ijpara.2016.05.008. Epub 2016 Jul 2. Int J Parasitol. 2017. PMID: 27381764 Free PMC article. Review.
Cited by
-
Population genomic evidence of Plasmodium vivax Southeast Asian origin.Sci Adv. 2021 Apr 28;7(18):eabc3713. doi: 10.1126/sciadv.abc3713. Print 2021 Apr. Sci Adv. 2021. PMID: 33910900 Free PMC article.
-
Why Plasmodium vivax and Plasmodium falciparum are so different? A tale of two clades and their species diversities.Malar J. 2022 May 3;21(1):139. doi: 10.1186/s12936-022-04130-9. Malar J. 2022. PMID: 35505356 Free PMC article. Review.
-
Evaluation of Histidine-Rich Proteins 2 and 3 Gene Deletions in Plasmodium falciparum in Endemic Areas of the Brazilian Amazon.Int J Environ Res Public Health. 2020 Dec 26;18(1):123. doi: 10.3390/ijerph18010123. Int J Environ Res Public Health. 2020. PMID: 33375379 Free PMC article.
-
Plasmodium vivax Malaria Viewed through the Lens of an Eradicated European Strain.Mol Biol Evol. 2020 Mar 1;37(3):773-785. doi: 10.1093/molbev/msz264. Mol Biol Evol. 2020. PMID: 31697387 Free PMC article.
-
Contribution of Travelers to Plasmodium Vivax Malaria in South West Delhi, India: Cross-Sectional Survey.JMIR Public Health Surveill. 2025 Jan 8;11:e50058. doi: 10.2196/50058. JMIR Public Health Surveill. 2025. PMID: 39780394 Free PMC article.
References
-
- McNeill, W. H. Plagues and Peoples (Anchor Press/Doubleday, 1976).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

