Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling
- PMID: 29386531
- PMCID: PMC5792481
- DOI: 10.1038/s41598-018-20114-9
Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling
Abstract
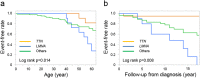
Dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM) are genetically and phenotypically heterogeneous. Cardiac function is improved after treatment in some cardiomyopathy patients, but little is known about genetic predictors of long-term outcomes and myocardial recovery following medical treatment. To elucidate the genetic basis of cardiomyopathy in Japan and the genotypes involved in prognosis and left ventricular reverse remodeling (LVRR), we performed targeted sequencing on 120 DCM (70 sporadic and 50 familial) and 52 HCM (15 sporadic and 37 familial) patients and integrated their genotypes with clinical phenotypes. Among the 120 DCM patients, 20 (16.7%) had TTN truncating variants and 13 (10.8%) had LMNA variants. TTN truncating variants were the major cause of sporadic DCM (21.4% of sporadic cases) as with Caucasians, whereas LMNA variants, which include a novel recurrent LMNA E115M variant, were the most frequent in familial DCM (24.0% of familial cases) unlike Caucasians. Of the 52 HCM patients, MYH7 and MYBPC3 variants were the most common (12 (23.1%) had MYH7 variants and 11 (21.2%) had MYBPC3 variants) as with Caucasians. DCM patients harboring TTN truncating variants had better prognosis than those with LMNA variants. Most patients with TTN truncating variants achieved LVRR, unlike most patients with LMNA variants.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Maron BJ. Hypertrophic cardiomyopathy: a systematic review. Jama. 2002;287:1308–1320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

