Genome-wide associations identify novel candidate loci associated with genetic susceptibility to tuberculosis in wild boar
- PMID: 29386541
- PMCID: PMC5792637
- DOI: 10.1038/s41598-018-20158-x
Genome-wide associations identify novel candidate loci associated with genetic susceptibility to tuberculosis in wild boar
Abstract
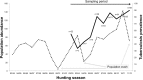
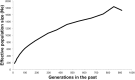
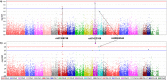
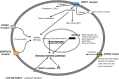
Tuberculosis (TB) affects a wide range of host species worldwide. Understanding host-pathogen co-evolution remains a global challenge owing to complex interactions among host genetic factors, pathogen traits and environmental conditions. We used an endemic wild boar population that had undergone a huge increase in Mycobacterium bovis infection prevalence, from 45% in 2002/06 to 83% in 2009/12, to understand the effects of host genetics on host TB outcomes and disease dynamics. Host genomic variation was characterized using a high-density single nucleotide polymorphism (SNP) array, while host TB phenotype was assessed using both gross pathology and mycobacterial culture. Two complementary genome-wide association (GWAS) analyses were conducted: (i) infected-uninfected; and (ii) 2002/06-2009/12. The SNPs with the highest allelic frequency differences between time-periods and TB outcomes were identified and validated in a large dataset. In addition, we quantified the expression levels of some of their closest genes. These analyses highlighted various SNPs (i.e. rs81465339, rs81394585, rs81423166) and some of the closest genes (i.e. LOC102164072, BDNF/NT-3, NTRK2, CDH8, IGSF21) as candidates for host genetic susceptibility. In addition to TB-driven selection, our findings outline the putative role of demographic events in shaping genomic variation in natural populations and how population crashes and drift may impact host genetic susceptibility to TB over time.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
[Frontier of mycobacterium research--host vs. mycobacterium].Kekkaku. 2005 Sep;80(9):613-29. Kekkaku. 2005. PMID: 16245793 Japanese.
-
Pathogen lineage-based genome-wide association study identified CD53 as susceptible locus in tuberculosis.J Hum Genet. 2017 Dec;62(12):1015-1022. doi: 10.1038/jhg.2017.82. Epub 2017 Sep 7. J Hum Genet. 2017. PMID: 28878339 Free PMC article.
-
Tuberculosis, genetic diversity and fitness in the red deer, Cervus elaphus.Infect Genet Evol. 2016 Sep;43:203-12. doi: 10.1016/j.meegid.2016.05.031. Epub 2016 May 28. Infect Genet Evol. 2016. PMID: 27245150
-
Evidence of the role of European wild boar as a reservoir of Mycobacterium tuberculosis complex.Vet Microbiol. 2008 Feb 5;127(1-2):1-9. doi: 10.1016/j.vetmic.2007.10.002. Epub 2007 Oct 10. Vet Microbiol. 2008. PMID: 18023299 Review.
-
The Role of Host Genetics (and Genomics) in Tuberculosis.Microbiol Spectr. 2016 Oct;4(5). doi: 10.1128/microbiolspec.TBTB2-0011-2016. Microbiol Spectr. 2016. PMID: 27787193 Review.
Cited by
-
Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary.Mol Ecol Resour. 2019 Jul;19(4):1015-1026. doi: 10.1111/1755-0998.13020. Epub 2019 May 17. Mol Ecol Resour. 2019. PMID: 30972949 Free PMC article.
-
Genomic regions associated with pseudorabies virus infection status in naturally infected feral swine (Sus scrofa).Front Genet. 2023 Nov 23;14:1292671. doi: 10.3389/fgene.2023.1292671. eCollection 2023. Front Genet. 2023. PMID: 38075681 Free PMC article.
-
A Study of the Resistance of Hu Sheep Lambs to Escherichia coli F17 Based on Whole Genome Sequencing.Animals (Basel). 2024 Jan 3;14(1):161. doi: 10.3390/ani14010161. Animals (Basel). 2024. PMID: 38200892 Free PMC article.
-
Identification of candidate genes associated with bacterial and viral infections in wild boars hunted in Tuscany (Italy).Sci Rep. 2022 May 17;12(1):8145. doi: 10.1038/s41598-022-12353-8. Sci Rep. 2022. PMID: 35581286 Free PMC article.
-
Experimental Pulmonary Tuberculosis in the Absence of Detectable Brain Infection Induces Neuroinflammation and Behavioural Abnormalities in Male BALB/c Mice.Int J Mol Sci. 2020 Dec 13;21(24):9483. doi: 10.3390/ijms21249483. Int J Mol Sci. 2020. PMID: 33322180 Free PMC article.
References
-
- Gortázar C, et al. The status of tuberculosis in European wild mammals. Mamm. Rev. 2012;42:193–206. doi: 10.1111/j.1365-2907.2011.00191.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

