Meta-analysis of transcriptomic changes in optic nerve injury and neurodegenerative models reveals a fundamental response to injury throughout the central nervous system
- PMID: 29386873
- PMCID: PMC5757855
Meta-analysis of transcriptomic changes in optic nerve injury and neurodegenerative models reveals a fundamental response to injury throughout the central nervous system
Abstract
Purpose: Injury to the central nervous system (CNS) leads to transcriptional changes that effect tissue function and govern the process of neurodegeneration. Numerous microarray and RNA-Seq studies have been performed to identify these transcriptional changes in the retina following optic nerve injury and elsewhere in the CNS following a variety of insults. We reasoned that conserved transcriptional changes between injury paradigms would be important contributors to the neurodegenerative process. Therefore, we compared the expression results from heterogeneous studies of optic nerve injury and neurodegenerative models.
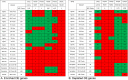
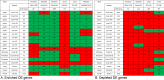
Methods: Expression data was collected from the Gene Expression Omnibus. A uniform method for normalizing expression data and detecting differentially expressed (DE) genes was used to compare the transcriptomes from models of acute optic nerve injury (AONI), chronic optic nerve injury (CONI) and brain neurodegeneration. DE genes were split into genes that were more or less prevalent in the injured condition than the control condition (enriched and depleted, respectively) and transformed into their human orthologs so that transcriptomes from different species could be compared. Biologic significance of shared genes was assessed by analyzing lists of shared genes for gene ontology (GO) term over-representation and for representation in KEGG pathways.
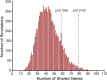
Results: There was significant overlap of enriched DE genes between transcriptomes of AONI, CONI and neurodegeneration studies even though the overall concordance between datasets was low. The depleted DE genes identified between AONI and CONI models were significantly overlapping, but this significance did not extend to comparisons between optic nerve injury models and neurodegeneration studies. The GO terms overrepresented among the enriched genes shared between AONI, CONI and neurodegeneration studies were related to innate immune processes like the complement system and interferon signaling. KEGG pathway analysis revealed that transcriptional alteration between JAK-STAT, PI3K-AKT and TNF signaling, among others, were conserved between all models that were analyzed.
Conclusions: There is a conserved transcriptional response to injury in the CNS. This transcriptional response is driven by the activation of the innate immune system and several regulatory pathways. Understanding the cellular origin of these pathways and the pathological consequences of their activation is essential for understanding and treating neurodegenerative disease.
Figures


References
-
- Herdegen T, Skene P, Bähr M. The c-Jun transcription factor – bipotential mediator of neuronal death, survival and regeneration. Trends Neurosci. 1997;20:227–31. - PubMed
-
- Li Y, Schlamp CL, Nickells RW. Experimental induction of retinal ganglion cell death in adult mice. Invest Ophthalmol Vis Sci. 1999;40:1004–8. - PubMed
-
- Villegas-Pérez M-P, Vidal-Sanz M, Rasminsky M, Bray GM, Aguayo AJ. Rapid and protracted phases of retinal ganglion cell loss follow axotomy in the optic nerve of adult rats. J Neurobiol. 1993;24:23–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

