Abnormal Ergosterol Biosynthesis Activates Transcriptional Responses to Antifungal Azoles
- PMID: 29387050
- PMCID: PMC5776110
- DOI: 10.3389/fmicb.2018.00009
Abnormal Ergosterol Biosynthesis Activates Transcriptional Responses to Antifungal Azoles
Abstract
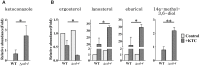
Fungi transcriptionally upregulate expression of azole efflux pumps and ergosterol biosynthesis pathway genes when exposed to antifungal agents that target ergosterol biosynthesis. To date, these transcriptional responses have been shown to be dependent on the presence of the azoles and/or depletion of ergosterol. Using an inducible promoter to regulate Neurospora crassa erg11, which encodes the major azole target, sterol 14α-demethylase, we were able to demonstrate that the CDR4 azole efflux pump can be transcriptionally activated by ergosterol biosynthesis inhibition even in the absence of azoles. By analyzing ergosterol deficient mutants, we demonstrate that the transcriptional responses by cdr4 and, unexpectedly, genes encoding ergosterol biosynthesis enzymes (erg genes) that are responsive to azoles, are not dependent on ergosterol depletion. Nonetheless, deletion of erg2, which encodes C-8 sterol isomerase, also induced expression of cdr4. Deletion of erg2 also induced the expression of erg24, the gene encoding C-14 sterol reductase, but not other tested erg genes which were responsive to erg11 inactivation. This indicates that inhibition of specific steps of ergosterol biosynthesis can result in different transcriptional responses, which is further supported by our results obtained using different ergosterol biosynthesis inhibitors. Together with the sterol profiles, these results suggest that the transcriptional responses by cdr4 and erg genes are associated with accumulation of specific sterol intermediate(s). This was further supported by the fact that when the erg2 mutant was treated with ketoconazole, upstream inhibition overrode the effects by downstream inhibition on ergosterol biosynthesis pathway. Even though cdr4 expression is associated with the accumulation of sterol intermediates, intra- and extracellular sterol analysis by HPLC-MS indicated that the transcriptional induction of cdr4 did not result in efflux of the accumulated intermediate(s). This study demonstrates, by detailed genetic and chemical analysis, that transcriptional responses by a major efflux pump and genes of the ergosterol biosynthesis pathway to ergosterol biosynthesis inhibitors can be independent of the presence of the drugs and are linked with the accumulation of ergosterol intermediate(s).
Keywords: C-8 sterol isomerase; azoles; efflux pump; sterol 14α-demethylase; sterol intermediate; stress response; tcu-1 promoter.
Figures





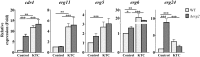

References
-
- Agarwal A. K., Rogers P. D., Baerson S. R., Jacob M. R., Barker K. S., Cleary J. D., et al. (2003). Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278 34998–35015. 10.1074/jbc.M306291200 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

