Plant Deubiquitinases and Their Role in the Control of Gene Expression Through Modification of Histones
- PMID: 29387079
- PMCID: PMC5776116
- DOI: 10.3389/fpls.2017.02274
Plant Deubiquitinases and Their Role in the Control of Gene Expression Through Modification of Histones
Abstract
Selective degradation of proteins in the cell occurs through ubiquitination, which consists of post-translational deposition of ubiquitin on proteins to target them for degradation by proteases. However, ubiquitination does not only impact on protein stability, but promotes changes in their functions. Whereas the deposition of ubiquitin has been amply studied and discussed, the antagonistic activity, deubiquitination, is just emerging and the full model and players involved in this mechanism are far from being completely understood. Nevertheless, it is the dynamic balance between ubiquitination and deubiquitination that is essential for the development and homeostasis of organisms. In this review, we present a detailed analysis of the members of the deubiquitinase (DUB) superfamily in plants and its division in different clades. We describe current knowledge in the molecular and functional characterisation of DUB proteins, focusing primarily on Arabidopsis thaliana. In addition, the striking function of the duality between ubiquitination and deubiquitination in the control of gene expression through the modification of chromatin is discussed and, using the available information of the activities of the DUB superfamily in yeast and animals as scaffold, we propose possible scenarios for the role of these proteins in plants.
Keywords: Arabidopsis; chromatin; deubiquitination; epigenetics; gene expression; histone modifications; plant development; ubiquitin.
Figures

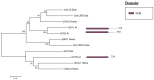




References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

