Bioinformatics analysis of RNA sequencing data reveals multiple key genes in uterine corpus endometrial carcinoma
- PMID: 29387216
- PMCID: PMC5769370
- DOI: 10.3892/ol.2017.7346
Bioinformatics analysis of RNA sequencing data reveals multiple key genes in uterine corpus endometrial carcinoma
Abstract
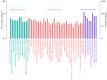
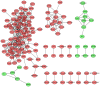
In the present study, the RNA sequencing (RNA-seq) data of uterine corpus endometrial carcinoma (UCEC) samples were collected and analyzed using bioinformatics tools to identify potential genes associated with the development of UCEC. UCEC RNA-seq data were downloaded from The Cancer Genome Atlas database. Differential analysis was performed using edgeR software. A false discovery rate <0.01 and |log2(fold change)|>1 were set as the cut-off criteria to screen for differentially expressed genes (DEGs). Differential gene co-expression analysis was performed using R/EBcoexpress package in R. DEGs in the gene co-expression network were subjected to Gene Ontology analysis using the Database for Annotation, Visualization and Integration Discovery. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis was also performed on the DEGs using KOBAS 2.0 software. The ConnectivityMap database was used to identify novel drug candidates. A total of 3,742 DEGs were identified among the 552 UCEC samples and 35 normal controls, and comprised 2,580 upregulated and 1,162 downregulated genes. A gene co-expression network consisting of 129 DEGs and 368 edges was constructed. Genes were associated with the cell cycle and the tumor protein p53 signaling pathway. Three modules were identified, in which genes were associated with the mitotic cell cycle, nuclear division and the M phase of the mitotic cell cycle. Multiple key hub genes were identified, including cell division cycle 20, cyclin B2, non-SMC condensin I complex subunit H, BUB1 mitotic checkpoint serine/threonine kinase, cell division cycle associated 8, maternal embryonic leucine zipper kinase, MYB proto-oncogene like 2, TPX2, microtubule nucleation factor and non-SMC condensin I complex subunit G. In addition, the small molecule drug esculetin was implicated in the suppression of UCEC progression. Overall, the present study identified multiple key genes in UCEC and clinically relevant small molecule agents, thereby improving our understanding of UCEC and expanding perspectives on targeted therapy for this type of cancer.
Keywords: differentially expressed genes; gene co-expression network; small molecule drugs; uterine corpus endometrial carcinoma.
Figures


References
-
- Devis L, Moiola CP, Masia N, Martinez-Garcia E, Santacana M, Stirbat TV, Brochard-Wyart F, García Á, Alameda F, Cabrera S, et al. Activated leukocyte cell adhesion molecule (ALCAM) is a marker of recurrence and promotes cell migration, invasion, and metastasis in early-stage endometrioid endometrial cancer. J Pathol. 2017;241:475–487. doi: 10.1002/path.4851. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
