Opposing and Complementary Topographic Connectivity Gradients Revealed by Quantitative Analysis of Canonical and Noncanonical Hippocampal CA1 Inputs
- PMID: 29387780
- PMCID: PMC5790753
- DOI: 10.1523/ENEURO.0322-17.2018
Opposing and Complementary Topographic Connectivity Gradients Revealed by Quantitative Analysis of Canonical and Noncanonical Hippocampal CA1 Inputs
Abstract
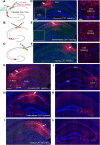
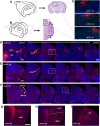
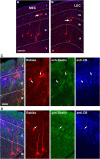
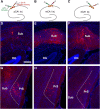
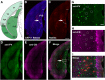
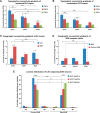
Physiological studies suggest spatial representation gradients along the CA1 proximodistal axis. To determine the underlying anatomical basis, we quantitatively mapped canonical and noncanonical inputs to excitatory neurons in dorsal hippocampal CA1 along the proximal-distal axis in mice of both sexes using monosynaptic rabies tracing. Our quantitative analyses show comparable strength of subiculum complex and entorhinal cortex (EC) inputs to CA1, significant inputs from presubiculum and parasubiculum to CA1, and a threefold stronger input to proximal versus distal CA1 from CA3. Noncanonical subicular complex inputs exhibit opposing topographic connectivity gradients whereby the subiculum-CA1 input strength systematically increases but the presubiculum-CA1 input strength decreases along the proximal-distal axis. The subiculum input strength cotracks that of the lateral EC, known to be less spatially selective than the medial EC. The functional significance of this organization is verified physiologically for subiculum-to-CA1 inputs. These results reveal a novel anatomical framework by which to determine the circuit bases for CA1 representations.
Keywords: Circuit connections; hippocampus; imaging; quantitative; viral tracing.
Figures


Similar articles
-
Hippocampal CA3 inhibitory neurons receive extensive noncanonical synaptic inputs from CA1 and subicular complex.J Comp Neurol. 2023 Sep;531(13):1333-1347. doi: 10.1002/cne.25510. Epub 2023 Jun 13. J Comp Neurol. 2023. PMID: 37312626 Free PMC article.
-
The entorhinal cortex of the monkey: VI. Organization of projections from the hippocampus, subiculum, presubiculum, and parasubiculum.J Comp Neurol. 2021 Mar;529(4):828-852. doi: 10.1002/cne.24983. Epub 2020 Aug 4. J Comp Neurol. 2021. PMID: 32656783 Free PMC article.
-
Noncanonical projections to the hippocampal CA3 regulate spatial learning and memory by modulating the feedforward hippocampal trisynaptic pathway.PLoS Biol. 2021 Dec 20;19(12):e3001127. doi: 10.1371/journal.pbio.3001127. eCollection 2021 Dec. PLoS Biol. 2021. PMID: 34928938 Free PMC article.
-
Noncanonical connections between the subiculum and hippocampal CA1.J Comp Neurol. 2016 Dec 1;524(17):3666-3673. doi: 10.1002/cne.24024. Epub 2016 May 6. J Comp Neurol. 2016. PMID: 27150503 Free PMC article. Review.
-
Functional diversity along the transverse axis of hippocampal area CA1.FEBS Lett. 2014 Aug 1;588(15):2470-6. doi: 10.1016/j.febslet.2014.06.004. Epub 2014 Jun 6. FEBS Lett. 2014. PMID: 24911200 Review.
Cited by
-
Subiculum as a generator of sharp wave-ripples in the rodent hippocampus.Cell Rep. 2021 Apr 20;35(3):109021. doi: 10.1016/j.celrep.2021.109021. Cell Rep. 2021. PMID: 33882307 Free PMC article.
-
Object-translocation induces event coding in the rat hippocampus.Commun Biol. 2025 May 24;8(1):797. doi: 10.1038/s42003-025-08241-2. Commun Biol. 2025. PMID: 40413308 Free PMC article.
-
Enhancing adult neurogenesis promotes contextual fear memory discrimination and activation of hippocampal-dorsolateral septal circuits.Behav Brain Res. 2021 Feb 5;399:112917. doi: 10.1016/j.bbr.2020.112917. Epub 2020 Sep 16. Behav Brain Res. 2021. PMID: 32949641 Free PMC article.
-
Whole-brain input mapping of the lateral versus medial anterodorsal bed nucleus of the stria terminalis in the mouse.Neurobiol Stress. 2023 Feb 15;23:100527. doi: 10.1016/j.ynstr.2023.100527. eCollection 2023 Mar. Neurobiol Stress. 2023. PMID: 36861029 Free PMC article.
-
Cell-specific switch for epileptiform activity: critical role of interneurons in the mouse subicular network.Cereb Cortex. 2023 May 9;33(10):6171-6183. doi: 10.1093/cercor/bhac493. Cereb Cortex. 2023. PMID: 36611229 Free PMC article.
References
-
- Amaral DG, Dolorfo C, Alvarez-Royo P (1991) Organization of CA1 projections to the subiculum: a PHA-L analysis in the rat. Hippocampus 1:415–435. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
