Involvement of the ubiquitin-proteasome system in the expression of extracellular matrix genes in retinal pigment epithelial cells
- PMID: 29387813
- PMCID: PMC5789218
- DOI: 10.1016/j.bbrep.2018.01.005
Involvement of the ubiquitin-proteasome system in the expression of extracellular matrix genes in retinal pigment epithelial cells
Abstract
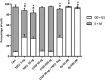
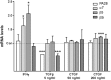
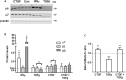
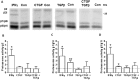
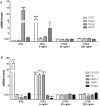
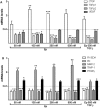
Emerging evidence suggests that dysfunction of the ubiquitin-proteasome system is involved in the pathogenesis of numerous senile degenerative diseases including retinal disorders. The aim of this study was to assess whether there is a link between proteasome regulation and retinal pigment epithelium (RPE)-mediated expression of extracellular matrix genes. For this purpose, human retinal pigment epithelial cells (ARPE-19) were treated with different concentrations of transforming growth factor-β (TGFβ), connective tissue growth factor (CTGF), interferon-γ (IFNγ) and the irreversible proteasome inhibitor epoxomicin. First, cytotoxicity and proliferation assays were carried out. The expression of proteasome-related genes and proteins was assessed and proteasome activity was determined. Then, expression of fibrosis-associated factors fibronectin (FN), fibronectin EDA domain (FN EDA), metalloproteinase-2 (MMP-2), tissue inhibitor of metalloproteinases-1 (TIMP-1) and peroxisome proliferator-associated receptor-γ (PPARγ) was assessed. The proteasome inhibitor epoxomicin strongly arrested cell cycle progression and down-regulated TGFβ gene expression, which in turn was shown to induce expression of pro-fibrogenic genes in ARPE-19 cells. Furthermore, epoxomicin induced a directional shift in the balance between MMP-2 and TIMP-1 and was associated with down-regulation of transcription of extracellular matrix genes FN and FN-EDA and up-regulation of the anti-fibrogenic factor PPARγ. In addition, both CTGF and TGFβ were shown to affect expression of proteasome-associated mRNA and protein levels. Our results suggest a link between proteasome activity and pro-fibrogenic mechanisms in the RPE, which could imply a role for proteasome-modulating agents in the treatment of retinal disorders characterized by RPE-mediated fibrogenic responses.
Keywords: AMD, age-related macular degeneration; ARPE-19, human retinal pigment epithelial cells; CNV, choroidal neovascularization; CTGF; CTGF, connective tissue growth factor; ECM, extracellular matrix; EMT, epithelial-mesenchymal transition; Epoxomicin; FN EDA, fibronectin EDA domain; FN, fibronectin; Fibrosis; IFNγ, interferon-γ; MMP-2, matrix metalloproteinase-2; PPARγ; PPARγ, peroxisome proliferator-associated receptor-γ; Proteasome; RPE; RPE, retinal pigment epithelium; Retina; TGFβ; TGFβ, transforming growth factor-β; TIMP-1, tissue inhibitor of metalloproteinases-1; UPS, ubiquitin-proteasome system; nAMD, neovascular age-related macular degeneration.
Figures
References
-
- Augood C.A., Vingerling J.R., De Jong P.T.V.M., Chakravarthy U., Seland J., Soubrane G., Tomazzoli L., Topouzis F., Bentham G., Rahu M., Vioque J., Young I.S., Fletcher A.E. Prevalence of age-related maculopathy in older Europeans - the European Eye Study (EUREYE) Arch. Ophthalmol. 2006;124:529–535. - PubMed
-
- Klein R., Chou C.F., Klein B.E.K., Zhang X.Z., Meuer S.M., Saaddine J.B. Prevalence of age-related macular degeneration in the US population. Arch. Ophthalmol. 2011;129:75–80. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

