Amino acid repeats avert mRNA folding through conservative substitutions and synonymous codons, regardless of codon bias
- PMID: 29387823
- PMCID: PMC5772840
- DOI: 10.1016/j.heliyon.2017.e00492
Amino acid repeats avert mRNA folding through conservative substitutions and synonymous codons, regardless of codon bias
Abstract
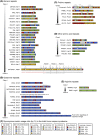
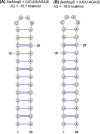
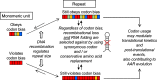
A significant number of proteins in all living species contains amino acid repeats (AARs) of various lengths and compositions, many of which play important roles in protein structure and function. Here, I have surveyed select homopolymeric single [(A)n] and double [(AB)n] AARs in the human proteome. A close examination of their codon pattern and analysis of RNA structure propensity led to the following set of empirical rules: (1) One class of amino acid repeats (Class I) uses a mixture of synonymous codons, some of which approximate the codon bias ratio in the overall human proteome; (2) The second class (Class II) disregards the codon bias ratio, and appears to have originated by simple repetition of the same codon (or just a few codons); and finally, (3) In all AARs (including Class I, Class II, and the in-betweens), the codons are chosen in a manner that precludes the formation of RNA secondary structure. It appears that the AAR genes have evolved by orchestrating a balance between codon usage and mRNA secondary structure. The insights gained here should provide a better understanding of AAR evolution and may assist in designing synthetic genes.
Keywords: Bioinformatics; Computational biology; Genetics; Structural biology.
Figures
Similar articles
-
Widespread position-specific conservation of synonymous rare codons within coding sequences.PLoS Comput Biol. 2017 May 5;13(5):e1005531. doi: 10.1371/journal.pcbi.1005531. eCollection 2017 May. PLoS Comput Biol. 2017. PMID: 28475588 Free PMC article.
-
The Yin and Yang of codon usage.Hum Mol Genet. 2016 Oct 1;25(R2):R77-R85. doi: 10.1093/hmg/ddw207. Epub 2016 Jun 27. Hum Mol Genet. 2016. PMID: 27354349 Free PMC article. Review.
-
Folding type specific secondary structure propensities of synonymous codons.IEEE Trans Nanobioscience. 2003 Sep;2(3):150-7. doi: 10.1109/tnb.2003.817024. IEEE Trans Nanobioscience. 2003. PMID: 15376949
-
[Influence of Synonymous Codon Bias on the RNA Secondary Structure in Influenza-A Viruses].Bing Du Xue Bao. 2016 Nov;32(6):773-81. Bing Du Xue Bao. 2016. PMID: 30004651 Chinese.
-
Decoding mechanisms by which silent codon changes influence protein biogenesis and function.Int J Biochem Cell Biol. 2015 Jul;64:58-74. doi: 10.1016/j.biocel.2015.03.011. Epub 2015 Mar 26. Int J Biochem Cell Biol. 2015. PMID: 25817479 Free PMC article. Review.
Cited by
-
Glutamine Codon Usage and polyQ Evolution in Primates Depend on the Q Stretch Length.Genome Biol Evol. 2018 Mar 1;10(3):816-825. doi: 10.1093/gbe/evy046. Genome Biol Evol. 2018. PMID: 29608721 Free PMC article.
-
A report on DNA sequence determinants in gene expression.Bioinformation. 2020 May 31;16(5):422-431. doi: 10.6026/97320630016422. eCollection 2020. Bioinformation. 2020. PMID: 32831525 Free PMC article.
-
Low Complexity Regions in Proteins and DNA are Poorly Correlated.Mol Biol Evol. 2023 Apr 4;40(4):msad084. doi: 10.1093/molbev/msad084. Mol Biol Evol. 2023. PMID: 37036379 Free PMC article.
-
The CpG Landscape of Protein Coding DNA in Vertebrates.Evol Appl. 2025 May 4;18(5):e70101. doi: 10.1111/eva.70101. eCollection 2025 May. Evol Appl. 2025. PMID: 40330995 Free PMC article.
-
Polyglutamine Repeats in Viruses.Mol Neurobiol. 2019 May;56(5):3664-3675. doi: 10.1007/s12035-018-1269-4. Epub 2018 Sep 4. Mol Neurobiol. 2019. PMID: 30182336 Free PMC article. Review.
References
-
- Mier P., Alanis-Lobato G., Andrade-Navarro M.A. Context characterization of amino acid homorepeats using evolution, position, and order. Proteins. 2017;85:709–719. - PubMed
-
- Andrade M.A., Perez-Iratxeta C., Ponting C.P. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 2001;134:117–131. - PubMed
-
- Yoshimura S.H., Hirano T. HEAT repeats – versatile arrays of amphiphilic helices working in crowded environments? J Cell Sci. 2016;129:3963–3970. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

