The curli regulator CsgD mediates stationary phase counter-silencing of csgBA in Salmonella Typhimurium
- PMID: 29388265
- PMCID: PMC5867262
- DOI: 10.1111/mmi.13919
The curli regulator CsgD mediates stationary phase counter-silencing of csgBA in Salmonella Typhimurium
Abstract
Integration of horizontally acquired genes into transcriptional networks is essential for the regulated expression of virulence in bacterial pathogens. In Salmonella enterica, expression of such genes is repressed by the nucleoid-associated protein H-NS, which recognizes and binds to AT-rich DNA. H-NS-mediated silencing must be countered by other DNA-binding proteins to allow expression under appropriate conditions. Some genes that can be transcribed by RNA polymerase (RNAP) associated with the alternative sigma factor σS or the housekeeping sigma factor σ70 in vitro appear to be preferentially transcribed by σS in the presence of H-NS, suggesting that σS may act as a counter-silencer. To determine whether σS directly counters H-NS-mediated silencing and whether co-regulation by H-NS accounts for the σS selectivity of certain promoters, we examined the csgBA operon, which is required for curli fimbriae expression and is known to be regulated by both H-NS and σS . Using genetics and in vitro biochemical analyses, we found that σS is not directly required for csgBA transcription, but rather up-regulates csgBA via an indirect upstream mechanism. Instead, the biofilm master regulator CsgD directly counter-silences the csgBA promoter by altering the DNA-protein complex structure to disrupt H-NS-mediated silencing in addition to directing the binding of RNAP.
© 2018 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

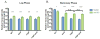
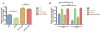



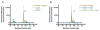
Similar articles
-
Relationships between H-NS, sigma S, SpvR and growth phase in the control of spvR, the regulatory gene of the Salmonella plasmid virulence operon.Mol Gen Genet. 1997 Oct;256(4):333-47. doi: 10.1007/s004380050577. Mol Gen Genet. 1997. PMID: 9393431
-
Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS.Mol Microbiol. 1994 Sep;13(6):1021-32. doi: 10.1111/j.1365-2958.1994.tb00493.x. Mol Microbiol. 1994. PMID: 7854117
-
MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium.Mol Microbiol. 2001 Jul;41(2):349-63. doi: 10.1046/j.1365-2958.2001.02529.x. Mol Microbiol. 2001. PMID: 11489123
-
The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium.Res Microbiol. 2003 Dec;154(10):659-67. doi: 10.1016/j.resmic.2003.08.005. Res Microbiol. 2003. PMID: 14643403 Review.
-
Control of virulence gene transcription by indirect readout in Vibrio cholerae and Salmonella enterica serovar Typhimurium.Environ Microbiol. 2017 Oct;19(10):3834-3845. doi: 10.1111/1462-2920.13838. Epub 2017 Jul 24. Environ Microbiol. 2017. PMID: 28631437 Free PMC article. Review.
Cited by
-
VirBR counter-silences HppX3 to promote conjugation of blaNDM-IncX3 plasmids.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf182. doi: 10.1093/nar/gkaf182. Nucleic Acids Res. 2025. PMID: 40103225 Free PMC article.
-
Refrigeration of eggs influences the virulence of Salmonella Typhimurium.Sci Rep. 2021 Sep 9;11(1):18026. doi: 10.1038/s41598-021-97135-4. Sci Rep. 2021. PMID: 34504138 Free PMC article.
-
Waking the neighbours: disruption of H-NS repression by overlapping transcription.Mol Microbiol. 2018 May;108(3):221-225. doi: 10.1111/mmi.13939. Epub 2018 Mar 23. Mol Microbiol. 2018. PMID: 29473964 Free PMC article.
-
MucR protein: Three decades of studies have led to the identification of a new H-NS-like protein.Mol Microbiol. 2025 Feb;123(2):154-167. doi: 10.1111/mmi.15261. Epub 2024 Apr 15. Mol Microbiol. 2025. PMID: 38619026 Review.
-
Designed α-sheet peptides disrupt uropathogenic E. coli biofilms rendering bacteria susceptible to antibiotics and immune cells.Sci Rep. 2023 Jun 7;13(1):9272. doi: 10.1038/s41598-023-36343-6. Sci Rep. 2023. PMID: 37286572 Free PMC article.
References
-
- Aparicio O, Geisberg JV, Sekinger E, Yang A, Moqtaderi Z, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol. 2005;Chapter 21(Unit 21.23) - PubMed
-
- Arnqvist A, Olsén A, Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

