Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project
- PMID: 29388947
- PMCID: PMC6520241
- DOI: 10.1038/gim.2017.241
Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project
Abstract
Purpose: Fresh-frozen (FF) tissue is the optimal source of DNA for whole-genome sequencing (WGS) of cancer patients. However, it is not always available, limiting the widespread application of WGS in clinical practice. We explored the viability of using formalin-fixed, paraffin-embedded (FFPE) tissues, available routinely for cancer patients, as a source of DNA for clinical WGS.
Methods: We conducted a prospective study using DNAs from matched FF, FFPE, and peripheral blood germ-line specimens collected from 52 cancer patients (156 samples) following routine diagnostic protocols. We compared somatic variants detected in FFPE and matching FF samples.
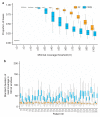
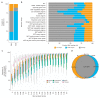
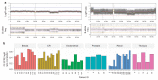
Results: We found the single-nucleotide variant agreement reached 71% across the genome and somatic copy-number alterations (CNAs) detection from FFPE samples was suboptimal (0.44 median correlation with FF) due to nonuniform coverage. CNA detection was improved significantly with lower reverse crosslinking temperature in FFPE DNA extraction (80 °C or 65 °C depending on the methods). Our final data showed somatic variant detection from FFPE for clinical decision making is possible. We detected 98% of clinically actionable variants (including 30/31 CNAs).
Conclusion: We present the first prospective WGS study of cancer patients using FFPE specimens collected in a routine clinical environment proving WGS can be applied in the clinic.
Keywords: clinical variant reporting; copy-number alteration; formalin-fixed, paraffin-embedded (FFPE); somatic variants; whole-genome sequencing.
Conflict of interest statement
J.B., M.H., M.T.R., Z.K., and D.R.B. are employees of Illumina, a public company that develops and markets systems for genetic analysis. The other authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

