Genome-Wide Identification and Expression Analysis of the HD-Zip Gene Family in Wheat (Triticum aestivum L.)
- PMID: 29389910
- PMCID: PMC5852566
- DOI: 10.3390/genes9020070
Genome-Wide Identification and Expression Analysis of the HD-Zip Gene Family in Wheat (Triticum aestivum L.)
Abstract
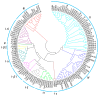
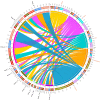
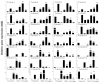
The homeodomain-leucine zipper (HD-Zip) gene family, as plant-specific transcription factors, plays an important role in plant development and growth as well as in the response to diverse stresses. Although HD-Zip genes have been extensively studied in many plants, they had not yet been studied in wheat, especially those involved in response to abiotic stresses. In this study, 46 wheat HD-Zip genes were identified using a genome-wide search method. Phylogenetic analysis classified these genes into four groups, numbered 4, 5, 17 and 20 respectively. In total, only three genes with A, B and D homoeologous copies were identified. Furthermore, the gene interaction networks found that the TaHDZ genes played a critical role in the regulatory pathway of organ development and osmotic stress. Finally, the expression profiles of the wheat HD-Zips in different tissues and under various abiotic stresses were investigated using the available RNA sequencing (RNA-Seq) data and then validated by quantitative real-time polymerase chain reaction (qRT-PCR) to obtain the tissue-specific and stress-responsive candidates. This study systematically identifies the HD-Zip gene family in wheat at the genome-wide level, providing important candidates for further functional analysis and contributing to the better understanding of the molecular basis of development and stress tolerance in wheat.
Keywords: HD-Zip; abiotic stress; expression profiles; gene family; wheat.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Characterization of wheat homeodomain-leucine zipper family genes and functional analysis of TaHDZ5-6A in drought tolerance in transgenic Arabidopsis.BMC Plant Biol. 2020 Jan 31;20(1):50. doi: 10.1186/s12870-020-2252-6. BMC Plant Biol. 2020. PMID: 32005165 Free PMC article.
-
Molecular interactions of the γ-clade homeodomain-leucine zipper class I transcription factors during the wheat response to water deficit.Plant Mol Biol. 2016 Mar;90(4-5):435-52. doi: 10.1007/s11103-015-0427-6. Epub 2016 Jan 23. Plant Mol Biol. 2016. PMID: 26803501
-
Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress.BMC Genomics. 2014 Nov 3;15:950. doi: 10.1186/1471-2164-15-950. BMC Genomics. 2014. PMID: 25362847 Free PMC article.
-
The roles of HD-ZIP proteins in plant abiotic stress tolerance.Front Plant Sci. 2022 Oct 12;13:1027071. doi: 10.3389/fpls.2022.1027071. eCollection 2022. Front Plant Sci. 2022. PMID: 36311122 Free PMC article. Review.
-
HD-ZIP Gene Family: Potential Roles in Improving Plant Growth and Regulating Stress-Responsive Mechanisms in Plants.Genes (Basel). 2021 Aug 17;12(8):1256. doi: 10.3390/genes12081256. Genes (Basel). 2021. PMID: 34440430 Free PMC article. Review.
Cited by
-
Genome-Wide Identification and Characterization of wALOG Family Genes Involved in Branch Meristem Development of Branching Head Wheat.Genes (Basel). 2018 Oct 19;9(10):510. doi: 10.3390/genes9100510. Genes (Basel). 2018. PMID: 30347757 Free PMC article.
-
Identification and Characterization of the HD-Zip Gene Family and Dimerization Analysis of HB7 and HB12 in Brassica napus L.Genes (Basel). 2022 Nov 17;13(11):2139. doi: 10.3390/genes13112139. Genes (Basel). 2022. PMID: 36421814 Free PMC article.
-
Analysis of the HD-Zip I transcription factor family in Salvia miltiorrhiza and functional research of SmHD-Zip12 in tanshinone synthesis.PeerJ. 2023 Jun 27;11:e15510. doi: 10.7717/peerj.15510. eCollection 2023. PeerJ. 2023. PMID: 37397009 Free PMC article.
-
Transcriptomic Profile of Tef (Eragrostis tef) in Response to Drought.Plants (Basel). 2024 Nov 2;13(21):3086. doi: 10.3390/plants13213086. Plants (Basel). 2024. PMID: 39520004 Free PMC article.
-
Plant Transcription Factors Involved in Drought and Associated Stresses.Int J Mol Sci. 2021 May 26;22(11):5662. doi: 10.3390/ijms22115662. Int J Mol Sci. 2021. PMID: 34073446 Free PMC article. Review.
References
-
- Wolfgang F., Phillips J., Salamini F., Bartels D. Two dehydration-inducible transcripts from the resurrection plant Craterostigma plantagineumencode interacting homeodomain-leucine zipper proteins. Plant J. 1998;15:413–421. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

