Comparative genomics reveals differences in mobile virulence genes of Escherichia coli O103 pathotypes of bovine fecal origin
- PMID: 29389941
- PMCID: PMC5794082
- DOI: 10.1371/journal.pone.0191362
Comparative genomics reveals differences in mobile virulence genes of Escherichia coli O103 pathotypes of bovine fecal origin
Abstract
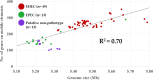
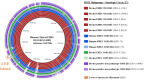
Escherichia coli O103, harbored in the hindgut and shed in the feces of cattle, can be enterohemorrhagic (EHEC), enteropathogenic (EPEC), or putative non-pathotype. The genetic diversity particularly that of virulence gene profiles within O103 serogroup is likely to be broad, considering the wide range in severity of illness. However, virulence descriptions of the E. coli O103 strains isolated from cattle feces have been primarily limited to major genes, such as Shiga toxin and intimin genes. Less is known about the frequency at which other virulence genes exist or about genes associated with the mobile genetic elements of E. coli O103 pathotypes. Our objective was to utilize whole genome sequencing (WGS) to identify and compare major and putative virulence genes of EHEC O103 (positive for Shiga toxin gene, stx1, and intimin gene, eae; n = 43), EPEC O103 (negative for stx1 and positive for eae; n = 13) and putative non-pathotype O103 strains (negative for stx and eae; n = 13) isolated from cattle feces. Six strains of EHEC O103 from human clinical cases were also included. All bovine EHEC strains (43/43) and a majority of EPEC (12/13) and putative non-pathotype strains (12/13) were O103:H2 serotype. Both bovine and human EHEC strains had significantly larger average genome sizes (P < 0.0001) and were positive for a higher number of adherence and toxin-based virulence genes and genes on mobile elements (prophages, transposable elements, and plasmids) than EPEC or putative non-pathotype strains. The genome size of the three pathotypes positively correlated (R2 = 0.7) with the number of genes carried on mobile genetic elements. Bovine strains clustered phylogenetically by pathotypes, which differed in several key virulence genes. The diversity of E. coli O103 pathotypes shed in cattle feces is likely reflective of the acquisition or loss of virulence genes carried on mobile genetic elements.
Conflict of interest statement
Figures

References
-
- Centers for Disease Control and Prevention (CDC). Shiga toxin-producing Escherichia coli (STEC) surveillance annual summary, 2012. Atlanta, Georgia: US Department of Health and Human Services, CDC, 2014.
-
- Abdullah UY, Al-Sultan II, Jassim HM, Ali YA, Khorsheed RM, Baig AA. Hemolytic uremic syndrome caused by Shiga toxin-producing Escherichia coli infections: an overview. Cloning & Transgenesis. 2014;3: 1–9.
-
- Ewers C, Janßen T, Wieler L. Avian pathogenic Escherichia coli (APEC). Berliner und Munchener tierarztliche Wochenschrift. 2002;116: 381–395. - PubMed
-
- Jordan DM, Cornick N, Torres AG, Dean-Nystrom EA, Kaper JB, Moon HW. Long polar fimbriae contribute to colonization by Escherichia coli O157: H7 in vivo. Infect Immun. 2004;72: 6168–6171. doi: 10.1128/IAI.72.10.6168-6171.2004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

