The Candida albicans stress response gene Stomatin-Like Protein 3 is implicated in ROS-induced apoptotic-like death of yeast phase cells
- PMID: 29389961
- PMCID: PMC5794166
- DOI: 10.1371/journal.pone.0192250
The Candida albicans stress response gene Stomatin-Like Protein 3 is implicated in ROS-induced apoptotic-like death of yeast phase cells
Abstract
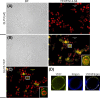
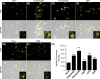
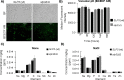
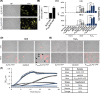
The ubiquitous presence of SPFH (Stomatin, Prohibitin, Flotillin, HflK/HflC) proteins in all domains of life suggests that their function would be conserved. However, SPFH functions are diverse with organism-specific attributes. SPFH proteins play critical roles in physiological processes such as mechanosensation and respiration. Here, we characterize the stomatin ORF19.7296/SLP3 in the opportunistic human pathogen Candida albicans. Consistent with the localization of stomatin proteins, a Slp3p-Yfp fusion protein formed visible puncta along the plasma membrane. We also visualized Slp3p within the vacuolar lumen. Slp3p primary sequence analyses identified four putative S-palmitoylation sites, which may facilitate membrane localization and are conserved features of stomatins. Plasma membrane insertion sequences are present in mammalian and nematode SPFH proteins, but are absent in Slp3p. Strikingly, Slp3p was present in yeast cells, but was absent in hyphal cells, thus categorizing it as a yeast-phase specific protein. Slp3p membrane fluorescence significantly increased in response to cellular stress caused by plasma membrane, cell wall, oxidative, or osmotic perturbants, implicating SLP3 as a general stress-response gene. A slp3Δ/Δ homozygous null mutant had no detected phenotype when slp3Δ/Δ mutants were grown in the presence of a variety of stress agents. Also, we did not observe a defect in ion accumulation, filamentation, endocytosis, vacuolar structure and function, cell wall structure, or cytoskeletal structure. However, SLP3 over-expression triggered apoptotic-like death following prolonged exposure to oxidative stress or when cells were induced to form hyphae. Our findings reveal the cellular localization of Slp3p, and for the first time associate Slp3p function with the oxidative stress response.
Conflict of interest statement
Figures



Similar articles
-
Mitochondrial targeting of Candida albicans SPFH proteins and requirement of stomatins for SDS-induced stress tolerance.Microbiol Spectr. 2025 Jan 7;13(1):e0173324. doi: 10.1128/spectrum.01733-24. Epub 2024 Dec 6. Microbiol Spectr. 2025. PMID: 39641539 Free PMC article.
-
Deletion analysis of LSm, FDF, and YjeF domains of Candida albicans Edc3 in hyphal growth and oxidative-stress response.J Microbiol. 2015 Feb;53(2):111-5. doi: 10.1007/s12275-015-4727-y. Epub 2015 Jan 28. J Microbiol. 2015. PMID: 25626365
-
The SPFH Protein Superfamily in Fungi: Impact on Mitochondrial Function and Implications in Virulence.Microorganisms. 2021 Nov 3;9(11):2287. doi: 10.3390/microorganisms9112287. Microorganisms. 2021. PMID: 34835412 Free PMC article. Review.
-
Mutational analysis of metacaspase CaMca1 and decapping activator Edc3 in the pathogenicity of Candida albicans.Fungal Genet Biol. 2016 Dec;97:18-23. doi: 10.1016/j.fgb.2016.10.007. Epub 2016 Nov 1. Fungal Genet Biol. 2016. PMID: 27815149
-
The lipid raft markers stomatin, prohibitin, flotillin, and HflK/C (SPFH)-domain proteins form an operon with NfeD proteins and function with apolar polyisoprenoid lipids.Crit Rev Microbiol. 2020 Feb;46(1):38-48. doi: 10.1080/1040841X.2020.1716682. Epub 2020 Jan 25. Crit Rev Microbiol. 2020. PMID: 31983249 Review.
Cited by
-
AFM-Based Correlative Microscopy Illuminates Human Pathogens.Front Cell Infect Microbiol. 2021 May 7;11:655501. doi: 10.3389/fcimb.2021.655501. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34026660 Free PMC article. Review.
-
Expression and clinical significance of SLP-2 in ovarian tumors.Oncol Lett. 2019 May;17(5):4626-4632. doi: 10.3892/ol.2019.10116. Epub 2019 Mar 5. Oncol Lett. 2019. PMID: 30944651 Free PMC article.
-
Triazine-Based Small Molecules: A Potential New Class of Compounds in the Antifungal Toolbox.Pathogens. 2023 Jan 12;12(1):126. doi: 10.3390/pathogens12010126. Pathogens. 2023. PMID: 36678474 Free PMC article.
-
Isobavachalcone exhibits antifungal and antibiofilm effects against C. albicans by disrupting cell wall/membrane integrity and inducing apoptosis and autophagy.Front Cell Infect Microbiol. 2024 Jan 22;14:1336773. doi: 10.3389/fcimb.2024.1336773. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38322671 Free PMC article.
-
An Amyloid Core Sequence in the Major Candida albicans Adhesin Als1p Mediates Cell-Cell Adhesion.mBio. 2019 Oct 8;10(5):e01766-19. doi: 10.1128/mBio.01766-19. mBio. 2019. PMID: 31594814 Free PMC article.
References
-
- Rivera-Milla E, Stuermer CA, Malaga-Trillo E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci. 2006;63(3):343–57. doi: 10.1007/s00018-005-5434-3 . - DOI - PMC - PubMed
-
- Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17(8):394–402. doi: 10.1016/j.tcb.2007.06.005 . - DOI - PubMed
-
- Lapatsina L, Brand J, Poole K, Daumke O, Lewin GR. Stomatin-domain proteins. Eur J Cell Biol. 2012;91(4):240–5. doi: 10.1016/j.ejcb.2011.01.018 . - DOI - PubMed
-
- Lee JH, Hsieh CF, Liu HW, Chen CY, Wu SC, Chen TW, et al. Lipid raft-associated stomatin enhances cell fusion. FASEB J. 2017;31(1):47–59. doi: 10.1096/fj.201600643R . - DOI - PubMed
-
- Reuter AT, Stuermer CA, Plattner H. Identification, localization, and functional implications of the microdomain-forming stomatin family in the ciliated protozoan Paramecium tetraurelia. Eukaryot Cell. 2013;12(4):529–44. doi: 10.1128/EC.00324-12 ; PubMed Central PMCID: PMCPMC3623435. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

