DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data
- PMID: 29391044
- PMCID: PMC5796597
- DOI: 10.1186/s40168-018-0401-z
DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data
Abstract
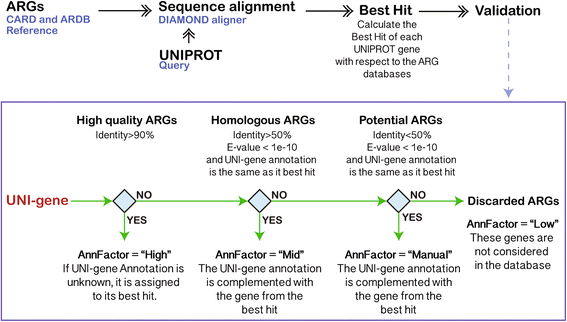
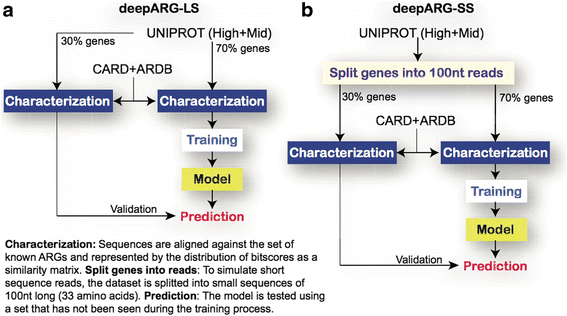
Background: Growing concerns about increasing rates of antibiotic resistance call for expanded and comprehensive global monitoring. Advancing methods for monitoring of environmental media (e.g., wastewater, agricultural waste, food, and water) is especially needed for identifying potential resources of novel antibiotic resistance genes (ARGs), hot spots for gene exchange, and as pathways for the spread of ARGs and human exposure. Next-generation sequencing now enables direct access and profiling of the total metagenomic DNA pool, where ARGs are typically identified or predicted based on the "best hits" of sequence searches against existing databases. Unfortunately, this approach produces a high rate of false negatives. To address such limitations, we propose here a deep learning approach, taking into account a dissimilarity matrix created using all known categories of ARGs. Two deep learning models, DeepARG-SS and DeepARG-LS, were constructed for short read sequences and full gene length sequences, respectively.
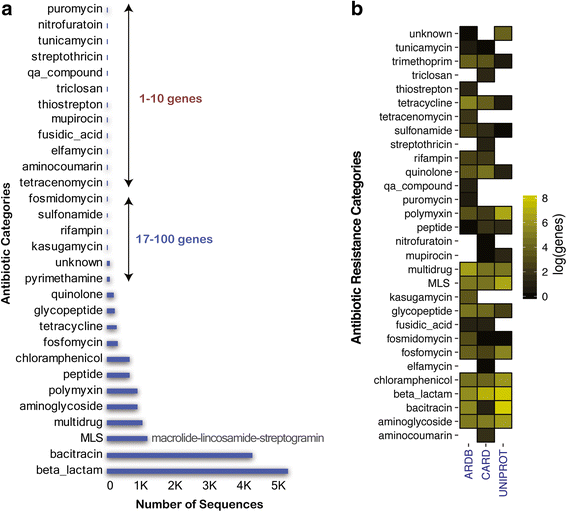
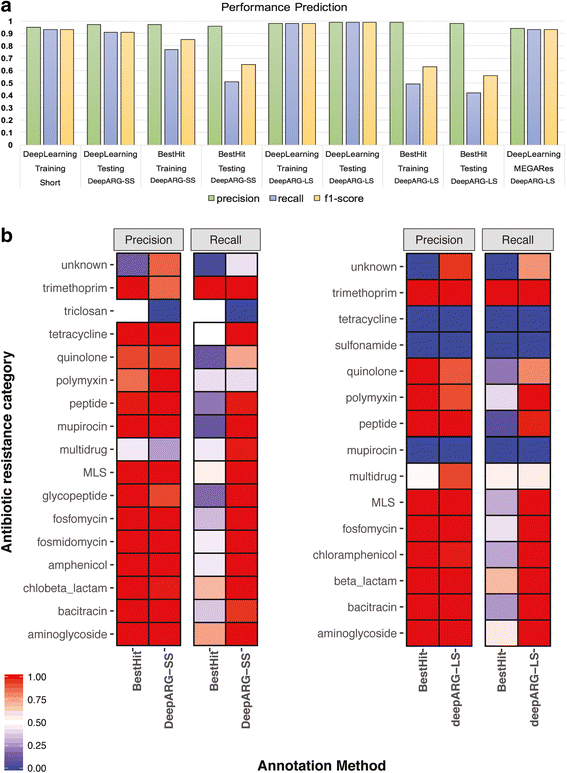
Results: Evaluation of the deep learning models over 30 antibiotic resistance categories demonstrates that the DeepARG models can predict ARGs with both high precision (> 0.97) and recall (> 0.90). The models displayed an advantage over the typical best hit approach, yielding consistently lower false negative rates and thus higher overall recall (> 0.9). As more data become available for under-represented ARG categories, the DeepARG models' performance can be expected to be further enhanced due to the nature of the underlying neural networks. Our newly developed ARG database, DeepARG-DB, encompasses ARGs predicted with a high degree of confidence and extensive manual inspection, greatly expanding current ARG repositories.
Conclusions: The deep learning models developed here offer more accurate antimicrobial resistance annotation relative to current bioinformatics practice. DeepARG does not require strict cutoffs, which enables identification of a much broader diversity of ARGs. The DeepARG models and database are available as a command line version and as a Web service at http://bench.cs.vt.edu/deeparg .
Keywords: Antibiotic resistance; Deep learning; Machine learning; Metagenomics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


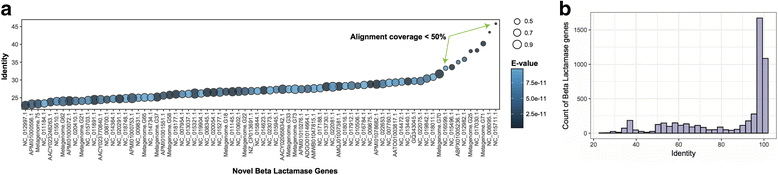
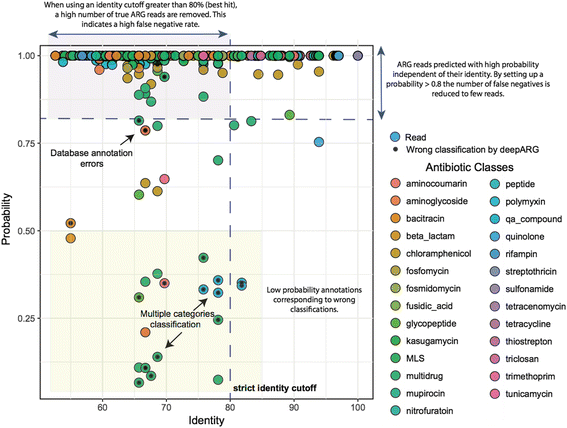

References
-
- O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. Rev Antimicrob Resist. 2016;1:1-84.
-
- O’Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Review on antimicrobial resistance. Rev Antimicrob Resist. 2014;
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

