Large-scale intact glycopeptide identification by Mascot database search
- PMID: 29391424
- PMCID: PMC5795011
- DOI: 10.1038/s41598-018-20331-2
Large-scale intact glycopeptide identification by Mascot database search
Erratum in
-
Author Correction: Large-scale intact glycopeptide identification by Mascot database search.Sci Rep. 2018 Jun 22;8(1):9771. doi: 10.1038/s41598-018-28147-w. Sci Rep. 2018. PMID: 29934554 Free PMC article.
Abstract
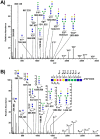
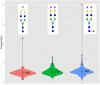
Workflows capable of determining glycopeptides in large-scale are missing in the field of glycoproteomics. We present an approach for automated annotation of intact glycopeptide mass spectra. The steps in adopting the Mascot search engine for intact glycopeptide analysis included: (i) assigning one letter codes for monosaccharides, (ii) linearizing glycan sequences and (iii) preparing custom glycoprotein databases. Automated annotation of both N- and O-linked glycopeptides was proven using standard glycoproteins. In a large-scale study, a total of 257 glycoproteins containing 970 unique glycosylation sites and 3447 non-redundant N-linked glycopeptide variants were identified in 24 serum samples. Thus, a single tool was developed that collectively allows the (i) elucidation of N- and O-linked glycopeptide spectra, (ii) matching glycopeptides to known protein sequences, and (iii) high-throughput, batch-wise analysis of large-scale glycoproteomics data sets.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

