Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes
- PMID: 29394990
- PMCID: PMC5985417
- DOI: 10.1016/j.ajhg.2017.12.015
Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes
Abstract
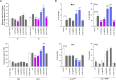
Exome sequencing has markedly enhanced the discovery of genes implicated in Mendelian disorders, particularly for individuals in whom a known clinical entity could not be assigned. This has led to the recognition that phenotypic heterogeneity resulting from allelic mutations occurs more commonly than previously appreciated. Here, we report that missense variants in CDC42, a gene encoding a small GTPase functioning as an intracellular signaling node, underlie a clinically heterogeneous group of phenotypes characterized by variable growth dysregulation, facial dysmorphism, and neurodevelopmental, immunological, and hematological anomalies, including a phenotype resembling Noonan syndrome, a developmental disorder caused by dysregulated RAS signaling. In silico, in vitro, and in vivo analyses demonstrate that mutations variably perturb CDC42 function by altering the switch between the active and inactive states of the GTPase and/or affecting CDC42 interaction with effectors, and differentially disturb cellular and developmental processes. These findings reveal the remarkably variable impact that dominantly acting CDC42 mutations have on cell function and development, creating challenges in syndrome definition, and exemplify the importance of functional profiling for syndrome recognition and delineation.
Keywords: Noonan syndrome; cardiac defects; developmental anomalies; exome sequencing; functional profiling; genotype-phenotype correlations; microcephaly; mutation spectrum; phenotypic heterogeneity; thrombocytopenia.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. - PubMed
-
- Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. - PMC - PubMed
-
- Menke L.A., van Belzen M.J., Alders M., Cristofoli F., Ehmke N., Fergelot P., Foster A., Gerkes E.H., Hoffer M.J., Horn D., DDD Study CREBBP mutations in individuals without Rubinstein-Taybi syndrome phenotype. Am. J. Med. Genet. A. 2016;170:2681–2693. - PubMed
-
- Lee C.S., Fu H., Baratang N., Rousseau J., Kumra H., Sutton V.R., Niceta M., Ciolfi A., Yamamoto G., Bertola D., Baylor-Hopkins Center for Mendelian Genomics Mutations in fibronectin cause a subtype of spondylometaphyseal dysplasia with “corner fractures”. Am. J. Hum. Genet. 2017;101:815–823. - PMC - PubMed
-
- Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A. Mutations impairing GSK3-mediated MAF phosphorylation cause cataract, deafness, intellectual disability, seizures, and a Down syndrome-like facies. Am. J. Hum. Genet. 2015;96:816–825. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

