Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus
- PMID: 29396526
- PMCID: PMC5797107
- DOI: 10.1038/s41598-018-20485-z
Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus
Abstract
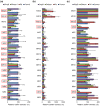
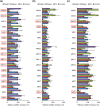
Staphylococcus aureus is a major causative agent for biofilm-associated infections. Inside biofilms, S. aureus cells are embedded in an extracellular matrix (ECM) composed of polysaccharide-intercellular adhesins (PIA), proteins, and/or extracellular DNA (eDNA). However, the importance of each component and the relationship among them in biofilms of diverse strains are largely unclear. Here, we characterised biofilms formed by 47 S. aureus clinical isolates. In most (42/47) of the strains, biofilm formation was augmented by glucose supplementation. Sodium chloride (NaCl)-triggered biofilm formation was more prevalent in methicillin-sensitive S. aureus (15/24) than in methicillin-resistant strain (1/23). DNase I most effectively inhibited and disrupted massive biofilms, and Proteinase K was also effective. Anti-biofilm effects of Dispersin B, which cleaves PIA, were restricted to PIA-dependent biofilms formed by specific strains and showed significant negative correlations with those of Proteinase K, suggesting independent roles of PIA and proteins in each biofilm. ECM profiling demonstrated that eDNA was present in all strains, although its level differed among strains and culture conditions. These results indicate that eDNA is the most common component in S. aureus biofilms, whereas PIA is important for a small number of isolates. Therefore, eDNA can be a primary target for developing eradication strategies against S. aureus biofilms.
Conflict of interest statement
Seiji Hori has received speaker’s honoraria from Daiichi Sankyo Co., Ltd. (Tokyo, Japan).
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

