Expression pattern of salt tolerance-related genes in Aegilops cylindrica
- PMID: 29398839
- PMCID: PMC5787114
- DOI: 10.1007/s12298-017-0483-2
Expression pattern of salt tolerance-related genes in Aegilops cylindrica
Abstract
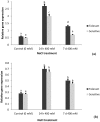
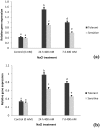
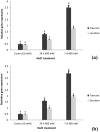
Aegilops cylindrica, a salt-tolerant gene pool of wheat, is a useful plant model for understanding mechanism of salt tolerance. A salt-tolerant USL26 and a salt-sensitive K44 genotypes of A. cylindrica, originating from Uremia Salt Lake shores in Northwest Iran and a non-saline Kurdestan province in West Iran, respectively, were identified based on screening evaluation and used for this work. The objective of the current study was to investigate the expression patterns of four genes related to ion homeostasis in this species. Under treatment of 400 mM NaCl, USL26 showed significantly higher root and shoot dry matter levels and K+ concentrations, together with lower Na+ concentrations than K44 genotype. A. cylindrica HKT1;5 (AecHKT1;5), SOS1 (AecSOS1), NHX1 (AecNHX1) and VP1 (AecVP1) were partially sequenced to design each gene specific primer. Quantitative real-time PCR showed a differential expression pattern of these genes between the two genotypes and between the root and shoot tissues. Expressions of AecHKT1;5 and AecSOS1 was greater in the roots than in the shoots of USL26 while AecNHX1 and AecVP1 were equally expressed in both tissues of USL26 and K44. The higher transcripts of AecHKT1;5 in the roots versus the shoots could explain both the lower Na+ in the shoots and the much lower Na+ and higher K+ concentrations in the roots/shoots of USL26 compared to K44. Therefore, the involvement of AecHKT1;5 in shoot-to-root handover of Na+ in possible combination with the exclusion of excessive Na+ from the root in the salt-tolerant genotype are suggested.
Keywords: HKT1;5; NHX1; SOS1; Salinity; Sodium exclusion.
Figures


References
-
- Arabbeigi M, Arzani A, Majidi MM, Kiani R, Sayed Tabatabaei BE, Habibi F. Salinity tolerance of Aegilops cylindrica genotypes collected from hyper-saline shores of Uremia Salt Lake using physiological traits and SSR markers. Acta Physiol Plant. 2014;36:2243–2251. doi: 10.1007/s11738-014-1602-0. - DOI
-
- Arzani A, Ashraf M. Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit Rev Plant Sci. 2016;35:146–189. doi: 10.1080/07352689.2016.1245056. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
