Sphingomyelin and GM1 Influence Huntingtin Binding to, Disruption of, and Aggregation on Lipid Membranes
- PMID: 29399649
- PMCID: PMC5793032
- DOI: 10.1021/acsomega.7b01472
Sphingomyelin and GM1 Influence Huntingtin Binding to, Disruption of, and Aggregation on Lipid Membranes
Abstract
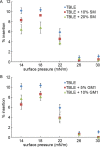
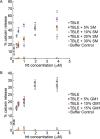
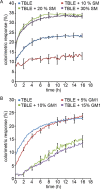
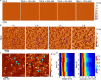
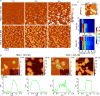
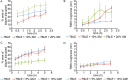
Huntington disease (HD) is an inherited neurodegenerative disease caused by the expansion beyond a critical threshold of a polyglutamine (polyQ) tract near the N-terminus of the huntingtin (htt) protein. Expanded polyQ promotes the formation of a variety of oligomeric and fibrillar aggregates of htt that accumulate into the hallmark proteinaceous inclusion bodies associated with HD. htt is also highly associated with numerous cellular and subcellular membranes that contain a variety of lipids. As lipid homeostasis and metabolism abnormalities are observed in HD patients, we investigated how varying both the sphingomyelin (SM) and ganglioside (GM1) contents modifies the interactions between htt and lipid membranes. SM composition is altered in HD, and GM1 has been shown to have protective effects in animal models of HD. A combination of Langmuir trough monolayer techniques, vesicle permeability and binding assays, and in situ atomic force microscopy (AFM) were used to directly monitor the interaction of a model, synthetic htt peptide and a full-length htt-exon1 recombinant protein with model membranes comprised of total brain lipid extract (TBLE) and varying amounts of exogenously added SM or GM1. The addition of either SM or GM1 decreased htt insertion into the lipid monolayers. However, TBLE vesicles with an increased SM content were more susceptible to htt-induced permeabilization, whereas GM1 had no effect on permeablization. Pure TBLE bilayers and TBLE bilayers enriched with GM1 developed regions of roughened, granular morphologies upon exposure to htt-exon1, but plateau-like domains with a smoother appearance formed in bilayers enriched with SM. Oligomeric aggregates were observed on all bilayer systems regardless of induced morphology. Collectively, these observations suggest that the lipid composition and its subsequent effects on membrane material properties strongly influence htt binding and aggregation on lipid membranes.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Gu X.; Greiner E. R.; Mishra R.; Kodali R.; Osmand A.; Finkbeiner S.; Steffan J. S.; Thompson L. M.; Wetzel R.; Yang X. W. Serines 13 and 16 Are Critical Determinants of Full-Length Human Mutant Huntingtin Induced Disease Pathogenesis in HD Mice. Neuron 2009, 64, 828–840. 10.1016/j.neuron.2009.11.020. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

