Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a Cretaceous Terrestrial Revolution
- PMID: 29399804
- PMCID: PMC6055841
- DOI: 10.1111/nph.15011
Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a Cretaceous Terrestrial Revolution
Abstract
Through the lens of the fossil record, angiosperm diversification precipitated a Cretaceous Terrestrial Revolution (KTR) in which pollinators, herbivores and predators underwent explosive co-diversification. Molecular dating studies imply that early angiosperm evolution is not documented in the fossil record. This mismatch remains controversial. We used a Bayesian molecular dating method to analyse a dataset of 83 genes from 644 taxa and 52 fossil calibrations to explore the effect of different interpretations of the fossil record, molecular clock models, data partitioning, among other factors, on angiosperm divergence time estimation. Controlling for different sources of uncertainty indicates that the timescale of angiosperm diversification is much less certain than previous molecular dating studies have suggested. Discord between molecular clock and purely fossil-based interpretations of angiosperm diversification may be a consequence of false precision on both sides. We reject a post-Jurassic origin of angiosperms, supporting the notion of a cryptic early history of angiosperms, but this history may be as much as 121 Myr, or as little as 23 Myr. These conclusions remain compatible with palaeobotanical evidence and a more general KTR in which major groups of angiosperms diverged later within the Cretaceous, alongside the diversification of pollinators, herbivores and their predators.
Keywords: Bayesian analysis; Cretaceous Terrestrial Revolution; angiosperms; divergence time; fossil record.
© 2018 The Authors. New Phytologist © 2018 New Phytologist Trust.
Figures
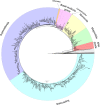


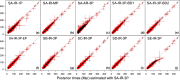


References
-
- Augusto L, Davies TJ, Delzon S, De Schrijver A. 2014. The enigma of the rise of angiosperms: can we untie the knot? Ecology Letters 17: 1326–1338. - PubMed
-
- Beaulieu JM, O'Meara BC, Crane P, Donoghue MJ. 2015. Heterogeneous rates of molecular evolution and diversification could explain the Triassic age estimate for angiosperms. Systematic Biology 64: 869–878. - PubMed
-
- Bell CD, Soltis DE, Soltis PS. 2005. The age of the angiosperms: a molecular timescale without a clock. Evolution 59: 1245–1258. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/G006660/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J00538X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N000609/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N000919/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

