Flex ddG: Rosetta Ensemble-Based Estimation of Changes in Protein-Protein Binding Affinity upon Mutation
- PMID: 29401388
- PMCID: PMC5980710
- DOI: 10.1021/acs.jpcb.7b11367
Flex ddG: Rosetta Ensemble-Based Estimation of Changes in Protein-Protein Binding Affinity upon Mutation
Abstract
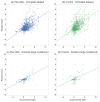
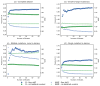
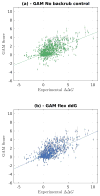
Computationally modeling changes in binding free energies upon mutation (interface ΔΔ G) allows large-scale prediction and perturbation of protein-protein interactions. Additionally, methods that consider and sample relevant conformational plasticity should be able to achieve higher prediction accuracy over methods that do not. To test this hypothesis, we developed a method within the Rosetta macromolecular modeling suite (flex ddG) that samples conformational diversity using "backrub" to generate an ensemble of models and then applies torsion minimization, side chain repacking, and averaging across this ensemble to estimate interface ΔΔ G values. We tested our method on a curated benchmark set of 1240 mutants, and found the method outperformed existing methods that sampled conformational space to a lesser degree. We observed considerable improvements with flex ddG over existing methods on the subset of small side chain to large side chain mutations, as well as for multiple simultaneous non-alanine mutations, stabilizing mutations, and mutations in antibody-antigen interfaces. Finally, we applied a generalized additive model (GAM) approach to the Rosetta energy function; the resulting nonlinear reweighting model improved the agreement with experimentally determined interface ΔΔ G values but also highlighted the necessity of future energy function improvements.
Figures


References
-
- Jubb HC, Pandurangan AP, Turner MA, Ochoa-Montaño B, Blundell TL, Ascher DB. Mutations at Protein-Protein Interfaces: Small Changes Over Big Surfaces Have Large Impacts on Human Health. Progress in Biophysics and Molecular Biology. 2017;128:3–13. doi: 10.1016/j.pbiomolbio.2016.10.002. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

