Development of a yeast internal-subunit eGFP labeling strategy and its application in subunit identification in eukaryotic group II chaperonin TRiC/CCT
- PMID: 29403048
- PMCID: PMC5799240
- DOI: 10.1038/s41598-017-18962-y
Development of a yeast internal-subunit eGFP labeling strategy and its application in subunit identification in eukaryotic group II chaperonin TRiC/CCT
Abstract
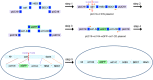
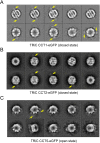
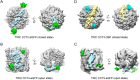
Unambiguous subunit assignment in a multicomponent complex is critical for thorough understanding of the machinery and its functionality. The eukaryotic group II chaperonin TRiC/CCT folds approximately 10% of cytosolic proteins and is important for the maintenance of cellular homeostasis. TRiC consists of two rings and each ring has eight homologous but distinct subunits. Unambiguous subunit identification of a macromolecular machine such as TRiC through intermediate or low-resolution cryo-EM map remains challenging. Here we present a yeast internal-subunit eGFP labeling strategy termed YISEL, which can quickly introduce an eGFP tag in the internal position of a target subunit by homologous recombination, and the tag labeled protein can be expressed in endogenous level. Through this method, the labeling efficiency and tag-occupancy is ensured, and the inserted tag is usually less mobile compared to that fused to the terminus. It can also be used to bio-engineer other tag in the internal position of a protein in yeast. By applying our YISEL strategy and combined with cryo-EM 3D reconstruction, we unambiguously identified all the subunits in the cryo-EM map of TRiC, demonstrating the potential for broad application of this strategy in accurate and efficient subunit identification in other challenging complexes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Yeast Inner-Subunit PA-NZ-1 Labeling Strategy for Accurate Subunit Identification in a Macromolecular Complex through Cryo-EM Analysis.J Mol Biol. 2018 May 11;430(10):1417-1425. doi: 10.1016/j.jmb.2018.03.026. Epub 2018 Apr 4. J Mol Biol. 2018. PMID: 29625202
-
4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.Proc Natl Acad Sci U S A. 2010 Mar 16;107(11):4967-72. doi: 10.1073/pnas.0913774107. Epub 2010 Mar 1. Proc Natl Acad Sci U S A. 2010. PMID: 20194787 Free PMC article.
-
Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.Nat Struct Mol Biol. 2016 Dec;23(12):1083-1091. doi: 10.1038/nsmb.3309. Epub 2016 Oct 24. Nat Struct Mol Biol. 2016. PMID: 27775711
-
TRiC/CCT Chaperonin: Structure and Function.Subcell Biochem. 2019;93:625-654. doi: 10.1007/978-3-030-28151-9_19. Subcell Biochem. 2019. PMID: 31939165 Review.
-
The ATP-powered gymnastics of TRiC/CCT: an asymmetric protein folding machine with a symmetric origin story.Curr Opin Struct Biol. 2019 Apr;55:50-58. doi: 10.1016/j.sbi.2019.03.002. Epub 2019 Apr 9. Curr Opin Struct Biol. 2019. PMID: 30978594 Free PMC article. Review.
Cited by
-
Advances in domain and subunit localization technology for electron microscopy.Biophys Rev. 2019 Apr;11(2):149-155. doi: 10.1007/s12551-019-00513-6. Epub 2019 Mar 5. Biophys Rev. 2019. PMID: 30834502 Free PMC article. Review.
-
Architecture and subunit arrangement of the complete Saccharomyces cerevisiae COMPASS complex.Sci Rep. 2018 Nov 27;8(1):17405. doi: 10.1038/s41598-018-35609-8. Sci Rep. 2018. PMID: 30479350 Free PMC article.
-
An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.Proc Natl Acad Sci U S A. 2019 Sep 24;116(39):19513-19522. doi: 10.1073/pnas.1903976116. Epub 2019 Sep 6. Proc Natl Acad Sci U S A. 2019. PMID: 31492816 Free PMC article.
-
The CCT chaperonin and actin modulate the ER and RNA-binding protein condensation during oogenesis and maintain translational repression of maternal mRNA and oocyte quality.Mol Biol Cell. 2024 Oct 1;35(10):ar131. doi: 10.1091/mbc.E24-05-0216. Epub 2024 Aug 21. Mol Biol Cell. 2024. PMID: 39167497 Free PMC article.
-
Lassa virus Z protein hijacks the autophagy machinery for efficient transportation by interrupting CCT2-mediated cytoskeleton network formation.Autophagy. 2024 Nov;20(11):2511-2528. doi: 10.1080/15548627.2024.2379099. Epub 2024 Jul 20. Autophagy. 2024. PMID: 39007910 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

