Lactobacillus rhamnosus GG Affects Microbiota and Suppresses Autophagy in the Intestines of Pigs Challenged with Salmonella Infantis
- PMID: 29403451
- PMCID: PMC5785727
- DOI: 10.3389/fmicb.2017.02705
Lactobacillus rhamnosus GG Affects Microbiota and Suppresses Autophagy in the Intestines of Pigs Challenged with Salmonella Infantis
Abstract
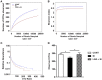
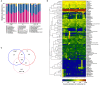
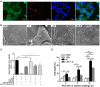
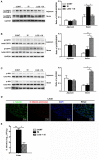
Salmonella enterica serovar Infantis (S. Infantis) is a common source of foodborne gastroenteritis worldwide. Here, Lactobacillus rhamnosus GG (LGG) was administrated to weaned piglets for 1 week before S. Infantis challenge. S. Infantis caused decreased ileal mucosal microbiota diversity, a dramatic Lactobacillus amylovorus bloom, and decreased abundance of Arsenicicoccus, Janibacter, Kocuria, Nocardioides, Devosia, Paracoccus, Psychrobacter, and Weissella. The beneficial effect of LGG correlated with the moderate expansion of L. amylovorus, L. agilis, and several members of the phyla Proteobacteria, Firmicutes, and Bacteroidetes. S. Infantis translocation to the liver was decreased in the LGG-pretreated piglets. An in vitro model of LGG and S. Infantis co-incubation (involving the porcine intestinal epithelial cell line IPEC-J2) was established, and nalidixic acid was used to kill the extracellular S. Infantis. LGG suppressed the initial S. Infantis invasion in the IPEC-J2 cells and deceased the rate of cell death. LGG inhibited S. Infantis-induced autophagy and promoted epidermal growth factor receptor (EGFR) and Akt phosphorylation in both the ileum and IPEC-J2 cells. Our findings suggest that LGG inhibited S. Infantis-induced autophagy by promoting EGFR-mediated activation of the negative mediator Akt, which, in turn, suppressed intestinal epithelial cell death and thus restricted systemic S. Infantis infection. LGG can restore the gut microbiota balance and preserve the autophagy-related intestinal epithelial barrier, thereby controlling infections.
Keywords: EGFR/Akt; Lactobacillus rhamnosus; Salmonella Infantis; autophagy; gut microbiota; pig.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

