Genomic and Phenotypic Variation in Morphogenetic Networks of Two Candida albicans Isolates Subtends Their Different Pathogenic Potential
- PMID: 29403478
- PMCID: PMC5780349
- DOI: 10.3389/fimmu.2017.01997
Genomic and Phenotypic Variation in Morphogenetic Networks of Two Candida albicans Isolates Subtends Their Different Pathogenic Potential
Abstract
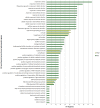
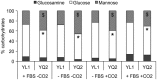
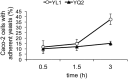
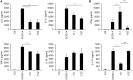
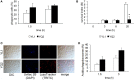
The transition from commensalism to pathogenicity of Candida albicans reflects both the host inability to mount specific immune responses and the microorganism's dimorphic switch efficiency. In this study, we used whole genome sequencing and microarray analysis to investigate the genomic determinants of the phenotypic changes observed in two C. albicans clinical isolates (YL1 and YQ2). In vitro experiments employing epithelial, microglial, and peripheral blood mononuclear cells were thus used to evaluate C. albicans isolates interaction with first line host defenses, measuring adhesion, susceptibility to phagocytosis, and induction of secretory responses. Moreover, a murine model of peritoneal infection was used to compare the in vivo pathogenic potential of the two isolates. Genome sequence and gene expression analysis of C. albicans YL1 and YQ2 showed significant changes in cellular pathways involved in environmental stress response, adhesion, filamentous growth, invasiveness, and dimorphic transition. This was in accordance with the observed marked phenotypic differences in biofilm production, dimorphic switch efficiency, cell adhesion, invasion, and survival to phagocyte-mediated host defenses. The mutations in key regulators of the hyphal growth pathway in the more virulent strain corresponded to an overall greater number of budding yeast cells released. Compared to YQ2, YL1 consistently showed enhanced pathogenic potential, since in vitro, it was less susceptible to ingestion by phagocytic cells and more efficient in invading epithelial cells, while in vivo YL1 was more effective than YQ2 in recruiting inflammatory cells, eliciting IL-1β response and eluding phagocytic cells. Overall, these results indicate an unexpected isolate-specific variation in pathways important for host invasion and colonization, showing how the genetic background of C. albicans may greatly affect its behavior both in vitro and in vivo. Based on this approach, we propose that the co-occurrence of changes in sequence and expression in genes and pathways driving dimorphic transition and pathogenicity reflects a selective balance between traits favoring dissemination of the pathogen and traits involved in host defense evasion. This study highlights the importance of investigating strain-level, rather than species level, differences, when determining fungal-host interactions and defining commensal or pathogen behavior.
Keywords: Candida albicans; biofilm; fungal isolates; genomic; host adaptation; pathogenic traits; phagocytes.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

