Using RNA Sequence and Structure for the Prediction of Riboswitch Aptamer: A Comprehensive Review of Available Software and Tools
- PMID: 29403526
- PMCID: PMC5780412
- DOI: 10.3389/fgene.2017.00231
Using RNA Sequence and Structure for the Prediction of Riboswitch Aptamer: A Comprehensive Review of Available Software and Tools
Abstract
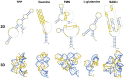
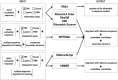
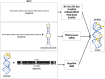
RNA molecules are essential players in many fundamental biological processes. Prokaryotes and eukaryotes have distinct RNA classes with specific structural features and functional roles. Computational prediction of protein structures is a research field in which high confidence three-dimensional protein models can be proposed based on the sequence alignment between target and templates. However, to date, only a few approaches have been developed for the computational prediction of RNA structures. Similar to proteins, RNA structures may be altered due to the interaction with various ligands, including proteins, other RNAs, and metabolites. A riboswitch is a molecular mechanism, found in the three kingdoms of life, in which the RNA structure is modified by the binding of a metabolite. It can regulate multiple gene expression mechanisms, such as transcription, translation initiation, and mRNA splicing and processing. Due to their nature, these entities also act on the regulation of gene expression and detection of small metabolites and have the potential to helping in the discovery of new classes of antimicrobial agents. In this review, we describe software and web servers currently available for riboswitch aptamer identification and secondary and tertiary structure prediction, including applications.
Keywords: RNA motif; RNA secondary structure; RNA tertiary structure; riboswitch; riboswitch aptamer prediction.
Figures

References
-
- Albert B., Bray D., Hopkin K. (2011). Fundamentos da Biologia Celular. Porto Alegre: Artmed.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

