X-ray and EPR Characterization of the Auxiliary Fe-S Clusters in the Radical SAM Enzyme PqqE
- PMID: 29405700
- PMCID: PMC5905707
- DOI: 10.1021/acs.biochem.7b01097
X-ray and EPR Characterization of the Auxiliary Fe-S Clusters in the Radical SAM Enzyme PqqE
Abstract
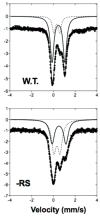
The Radical SAM (RS) enzyme PqqE catalyzes the first step in the biosynthesis of the bacterial cofactor pyrroloquinoline quinone, forming a new carbon-carbon bond between two side chains within the ribosomally synthesized peptide substrate PqqA. In addition to the active site RS 4Fe-4S cluster, PqqE is predicted to have two auxiliary Fe-S clusters, like the other members of the SPASM domain family. Here we identify these sites and examine their structure using a combination of X-ray crystallography and Mössbauer and electron paramagnetic resonance (EPR) spectroscopies. X-ray crystallography allows us to identify the ligands to each of the two auxiliary clusters at the C-terminal region of the protein. The auxiliary cluster nearest the RS site (AuxI) is in the form of a 2Fe-2S cluster ligated by four cysteines, an Fe-S center not seen previously in other SPASM domain proteins; this assignment is further supported by Mössbauer and EPR spectroscopies. The second, more remote cluster (AuxII) is a 4Fe-4S center that is ligated by three cysteine residues and one aspartate residue. In addition, we examined the roles these ligands play in catalysis by the RS and AuxII clusters using site-directed mutagenesis coupled with EPR spectroscopy. Lastly, we discuss the possible functional consequences that these unique AuxI and AuxII clusters may have in catalysis for PqqE and how these may extend to additional RS enzymes catalyzing the post-translational modification of ribosomally encoded peptides.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Structural Properties and Catalytic Implications of the SPASM Domain Iron-Sulfur Clusters in Methylorubrum extorquens PqqE.J Am Chem Soc. 2020 Jul 22;142(29):12620-12634. doi: 10.1021/jacs.0c02044. Epub 2020 Jul 9. J Am Chem Soc. 2020. PMID: 32643933 Free PMC article.
-
Electron Paramagnetic Resonance Spectroscopic Identification of the Fe-S Clusters in the SPASM Domain-Containing Radical SAM Enzyme PqqE.Biochemistry. 2019 Dec 24;58(51):5173-5187. doi: 10.1021/acs.biochem.9b00960. Epub 2019 Dec 11. Biochemistry. 2019. PMID: 31769977 Free PMC article.
-
Characterization of auxiliary iron-sulfur clusters in a radical S-adenosylmethionine enzyme PqqE from Methylobacterium extorquens AM1.FEBS Open Bio. 2017 Oct 18;7(12):1864-1879. doi: 10.1002/2211-5463.12314. eCollection 2017 Dec. FEBS Open Bio. 2017. PMID: 29226074 Free PMC article.
-
Mössbauer spectroscopy of Fe/S proteins.Biochim Biophys Acta. 2015 Jun;1853(6):1395-405. doi: 10.1016/j.bbamcr.2014.12.005. Epub 2014 Dec 10. Biochim Biophys Acta. 2015. PMID: 25498248 Review.
-
Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes.Biochim Biophys Acta. 2012 Nov;1824(11):1196-212. doi: 10.1016/j.bbapap.2012.07.009. Epub 2012 Jul 28. Biochim Biophys Acta. 2012. PMID: 22846545 Review.
Cited by
-
Structural Properties and Catalytic Implications of the SPASM Domain Iron-Sulfur Clusters in Methylorubrum extorquens PqqE.J Am Chem Soc. 2020 Jul 22;142(29):12620-12634. doi: 10.1021/jacs.0c02044. Epub 2020 Jul 9. J Am Chem Soc. 2020. PMID: 32643933 Free PMC article.
-
A Radical Clock Probe Uncouples H Atom Abstraction from Thioether Cross-Link Formation by the Radical S-Adenosyl-l-methionine Enzyme SkfB.Biochemistry. 2018 Aug 14;57(32):4816-4823. doi: 10.1021/acs.biochem.8b00537. Epub 2018 Jul 24. Biochemistry. 2018. PMID: 29965747 Free PMC article.
-
Biogenesis of the peptide-derived redox cofactor pyrroloquinoline quinone.Curr Opin Chem Biol. 2020 Dec;59:93-103. doi: 10.1016/j.cbpa.2020.05.001. Epub 2020 Jul 27. Curr Opin Chem Biol. 2020. PMID: 32731194 Free PMC article. Review.
-
Bioinformatic Mapping of Radical S-Adenosylmethionine-Dependent Ribosomally Synthesized and Post-Translationally Modified Peptides Identifies New Cα, Cβ, and Cγ-Linked Thioether-Containing Peptides.J Am Chem Soc. 2019 May 22;141(20):8228-8238. doi: 10.1021/jacs.9b01519. Epub 2019 May 13. J Am Chem Soc. 2019. PMID: 31059252 Free PMC article.
-
HygY Is a Twitch Radical SAM Epimerase with Latent Dehydrogenase Activity Revealed upon Mutation of a Single Cysteine Residue.J Am Chem Soc. 2021 Sep 22;143(37):15152-15158. doi: 10.1021/jacs.1c05727. Epub 2021 Sep 7. J Am Chem Soc. 2021. PMID: 34491039 Free PMC article.
References
-
- Duine JA. The PQQ story. J Biosci Bioeng. 1999;88:231–236. - PubMed
-
- Barr I, Latham JA, Iavarone AT, Chantarojsiri T, Hwang JD, Klinman JP. Demonstration That the Radical S-Adenosylmethionine (SAM) Enzyme PqqE Catalyzes de Novo Carbon-Carbon Cross-linking within a Peptide Substrate PqqA in the Presence of the Peptide Chaperone PqqD. J Biol Chem. 2016;291:8877–8884. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

