Active inference and the anatomy of oculomotion
- PMID: 29407941
- PMCID: PMC5884328
- DOI: 10.1016/j.neuropsychologia.2018.01.041
Active inference and the anatomy of oculomotion
Abstract
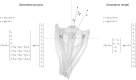
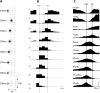
Given that eye movement control can be framed as an inferential process, how are the requisite forces generated to produce anticipated or desired fixation? Starting from a generative model based on simple Newtonian equations of motion, we derive a variational solution to this problem and illustrate the plausibility of its implementation in the oculomotor brainstem. We show, through simulation, that the Bayesian filtering equations that implement 'planning as inference' can generate both saccadic and smooth pursuit eye movements. Crucially, the associated message passing maps well onto the known connectivity and neuroanatomy of the brainstem - and the changes in these messages over time are strikingly similar to single unit recordings of neurons in the corresponding nuclei. Furthermore, we show that simulated lesions to axonal pathways reproduce eye movement patterns of neurological patients with damage to these tracts.
Keywords: Active inference; Brainstem; Free energy; Oculomotor; Predictive coding; Saccades.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures





References
-
- Anderson T.J., MacAskill M.R. Eye movements in patients with neurodegenerative disorders. Nat. Rev. Neurol. 2013;9(2):74–85. - PubMed
-
- Beal M.J. University of London United Kingdom; 2003. Variational Algorithms for Approximate Bayesian Inference.
-
- Berretta S., Bosco G., Giaquinta G., Smecca G., Perciavalle V. Cerebellar influences on accessory oculomotor nuclei of the rat: a neuroanatomical, immunohistochemical, and electrophysiological study. J. Comp. Neurol. 1993;338(1):50–66. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

