Phosphoproteomic analysis reveals Smad protein family activation following Rift Valley fever virus infection
- PMID: 29408900
- PMCID: PMC5800665
- DOI: 10.1371/journal.pone.0191983
Phosphoproteomic analysis reveals Smad protein family activation following Rift Valley fever virus infection
Erratum in
-
Correction: Phosphoproteomic analysis reveals Smad protein family activation following Rift Valley fever virus infection.PLoS One. 2018 Mar 15;13(3):e0194633. doi: 10.1371/journal.pone.0194633. eCollection 2018. PLoS One. 2018. PMID: 29543894 Free PMC article.
Abstract
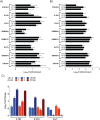
Rift Valley fever virus (RVFV) infects both ruminants and humans leading to a wide variance of pathologies dependent on host background and age. Utilizing a targeted reverse phase protein array (RPPA) to define changes in signaling cascades after in vitro infection of human cells with virulent and attenuated RVFV strains, we observed high phosphorylation of Smad transcription factors. This evolutionarily conserved family is phosphorylated by and transduces the activation of TGF-β superfamily receptors. Moreover, we observed that phosphorylation of Smad proteins required active RVFV replication and loss of NSs impaired this activation, further corroborating the RPPA results. Gene promoter analysis of transcripts altered after RVFV infection identified 913 genes that contained a Smad-response element. Functional annotation of these potential Smad-regulated genes clustered in axonal guidance, hepatic fibrosis and cell signaling pathways involved in cellular adhesion/migration, calcium influx, and cytoskeletal reorganization. Furthermore, chromatin immunoprecipitation confirmed the presence of a Smad complex on the interleukin 1 receptor type 2 (IL1R2) promoter, which acts as a decoy receptor for IL-1 activation.
Conflict of interest statement
Figures





References
-
- Smith H. The little-known determinants of virus pathogenicity. Scand J Infect Dis Suppl. 1980;Suppl 24:119–27. Epub 1980/01/01. . - PubMed
-
- Ly HJ, Ikegami T. Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins. Virol J. 2016;13:118 Epub 2016/07/03. doi: 10.1186/s12985-016-0573-8 . - DOI - PMC - PubMed
-
- Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, et al. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J Virol. 2001;75(3):1371–7. doi: 10.1128/JVI.75.3.1371-1377.2001 . - DOI - PMC - PubMed
-
- Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, et al. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS pathogens. 2008;4(1):e13 Epub 2008/01/30. doi: 10.1371/journal.ppat.0040013 . - DOI - PMC - PubMed
-
- Terasaki K, Ramirez SI, Makino S. Mechanistic Insight into the Host Transcription Inhibition Function of Rift Valley Fever Virus NSs and Its Importance in Virulence. PLoS neglected tropical diseases. 2016;10(10):e0005047 Epub 2016/10/07. doi: 10.1371/journal.pntd.0005047 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

