Effects of an EPSPS-transgenic soybean line ZUTS31 on root-associated bacterial communities during field growth
- PMID: 29408918
- PMCID: PMC5800644
- DOI: 10.1371/journal.pone.0192008
Effects of an EPSPS-transgenic soybean line ZUTS31 on root-associated bacterial communities during field growth
Abstract
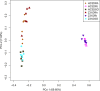
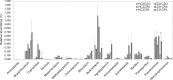
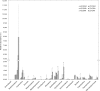
The increased worldwide commercial cultivation of transgenic crops during the past 20 years is accompanied with potential effects on the soil microbial communities, because many rhizosphere and endosphere bacteria play important roles in promoting plant health and growth. Previous studies reported that transgenic plants exert differential effects on soil microbial communities, especially rhizobacteria. Thus, this study compared the soybean root-associated bacterial communities between a 5-enolpyruvylshikimate-3-phosphate synthase -transgenic soybean line (ZUTS31 or simply Z31) and its recipient cultivar (Huachun3 or simply HC3) at the vegetative, flowering, and seed-filling stages. High-throughput sequencing of 16S rRNA gene (16S rDNA) V4 hypervariable region amplicons via Illumina MiSeq and real-time quantitative PCR (qPCR) were performed. Our results revealed no significant differences in the overall alpha diversity of root-associated bacterial communities at the three developmental stages and in the beta diversity of root-associated bacterial communities at the flowering stage between Z31 and HC3 under field growth. However, significant differences in the beta diversity of rhizosphere bacterial communities were found at the vegetative and seed-filling stages between the two groups. Furthermore, the results of next generation sequencing and qPCR showed that the relative abundances of root-associated main nitrogen-fixing bacterial genera, especially Bradyrhizobium in the roots, evidently changed from the flowering stage to the seed-filling stage. In conclusion, Z31 exerts transitory effects on the taxonomic diversity of rhizosphere bacterial communities at the vegetative and seed-filling stages compared to the control under field conditions. In addition, soybean developmental change evidently influences the main symbiotic nitrogen-fixing bacterial genera in the roots from the flowering stage to the seed-filling stage.
Conflict of interest statement
Figures




Similar articles
-
Impact of a G2-EPSPS & GAT Dual Transgenic Glyphosate-Resistant Soybean Line on the Soil Microbial Community under Field Conditions Affected by Glyphosate Application.Microbes Environ. 2020;35(4):ME20056. doi: 10.1264/jsme2.ME20056. Microbes Environ. 2020. PMID: 33162465 Free PMC article.
-
Enrichments/Derichments of Root-Associated Bacteria Related to Plant Growth and Nutrition Caused by the Growth of an EPSPS-Transgenic Maize Line in the Field.Front Microbiol. 2019 Jun 18;10:1335. doi: 10.3389/fmicb.2019.01335. eCollection 2019. Front Microbiol. 2019. PMID: 31275269 Free PMC article.
-
Impact of a Glyphosate-Tolerant Soybean Line on the Rhizobacteria, Revealed by Illumina MiSeq.J Microbiol Biotechnol. 2017 Mar 28;27(3):561-572. doi: 10.4014/jmb.1609.09008. J Microbiol Biotechnol. 2017. PMID: 27974727
-
Impact of Glyphosate on the Rhizosphere Microbial Communities of An EPSPS-Transgenic Soybean Line ZUTS31 by Metagenome Sequencing.Curr Genomics. 2018 Jan;19(1):36-49. doi: 10.2174/1389202918666170705162405. Curr Genomics. 2018. PMID: 29491731 Free PMC article.
-
The host niches of soybean rather than genetic modification or glyphosate application drive the assembly of root-associated microbial communities.Microb Biotechnol. 2022 Dec;15(12):2942-2957. doi: 10.1111/1751-7915.14164. Epub 2022 Nov 6. Microb Biotechnol. 2022. PMID: 36336802 Free PMC article.
Cited by
-
High-throughput sequencing analysis of microbial community diversity in response to indica and japonica bar-transgenic rice paddy soils.PLoS One. 2019 Sep 9;14(9):e0222191. doi: 10.1371/journal.pone.0222191. eCollection 2019. PLoS One. 2019. PMID: 31498816 Free PMC article.
-
Microbial Assemblages Associated with the Soil-Root Continuum of an Endangered Plant, Helianthemum songaricum Schrenk.Microbiol Spectr. 2023 Jun 15;11(3):e0338922. doi: 10.1128/spectrum.03389-22. Epub 2023 May 24. Microbiol Spectr. 2023. PMID: 37222598 Free PMC article.
-
Impact of a G2-EPSPS & GAT Dual Transgenic Glyphosate-Resistant Soybean Line on the Soil Microbial Community under Field Conditions Affected by Glyphosate Application.Microbes Environ. 2020;35(4):ME20056. doi: 10.1264/jsme2.ME20056. Microbes Environ. 2020. PMID: 33162465 Free PMC article.
-
Differential Impacts on Bacterial Composition and Abundance in Rhizosphere Compartments between Al-Tolerant and Al-Sensitive Soybean Genotypes in Acidic Soil.J Microbiol Biotechnol. 2020 Aug 28;30(8):1169-1179. doi: 10.4014/jmb.2003.03018. J Microbiol Biotechnol. 2020. PMID: 32522970 Free PMC article.
-
Effects of Insect-Resistant Maize HGK60 on Community Diversity of Bacteria and Fungi in Rhizosphere Soil.Plants (Basel). 2022 Oct 24;11(21):2824. doi: 10.3390/plants11212824. Plants (Basel). 2022. PMID: 36365278 Free PMC article.
References
-
- James C. Global Status of Commercialized Biotech/GM Crops: 2015. China Biotechnol; 2016; 36: 1–11. doi: 10.13523/j.cb.20160401 - DOI
-
- Benbrook CM. Trends in glyphosate herbicide use in the United States and globally. Environ Sci Eur; 2016; 28: 1–15. doi: 10.1186/s12302-015-0068-z - DOI - PMC - PubMed
-
- Dunfield KE, Germida JJ. Impact of genetically modified crops on soil- and plant-associated microbial communities. J Environ Qual; 2004; 33: 806–815. doi: 10.2134/jeq2004.0806 - DOI - PubMed
-
- Liu B, Zeng Q, Yan FM, Xu HG, Xu CR. Effects of transgenic plants on soil microorganisms. Plant Soil; 2005; 271: 1–13. doi: 10.1007/s11104-004-1610-8 - DOI
-
- Turrini A, Sbrana C, Giovannetti M. Belowground environmental effects of transgenic crops: a soil microbial perspective. Res Microbiol; 2015; 166: 121–131. doi: 10.1016/j.resmic.2015.02.006 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

