SCANPY: large-scale single-cell gene expression data analysis
- PMID: 29409532
- PMCID: PMC5802054
- DOI: 10.1186/s13059-017-1382-0
SCANPY: large-scale single-cell gene expression data analysis
Abstract
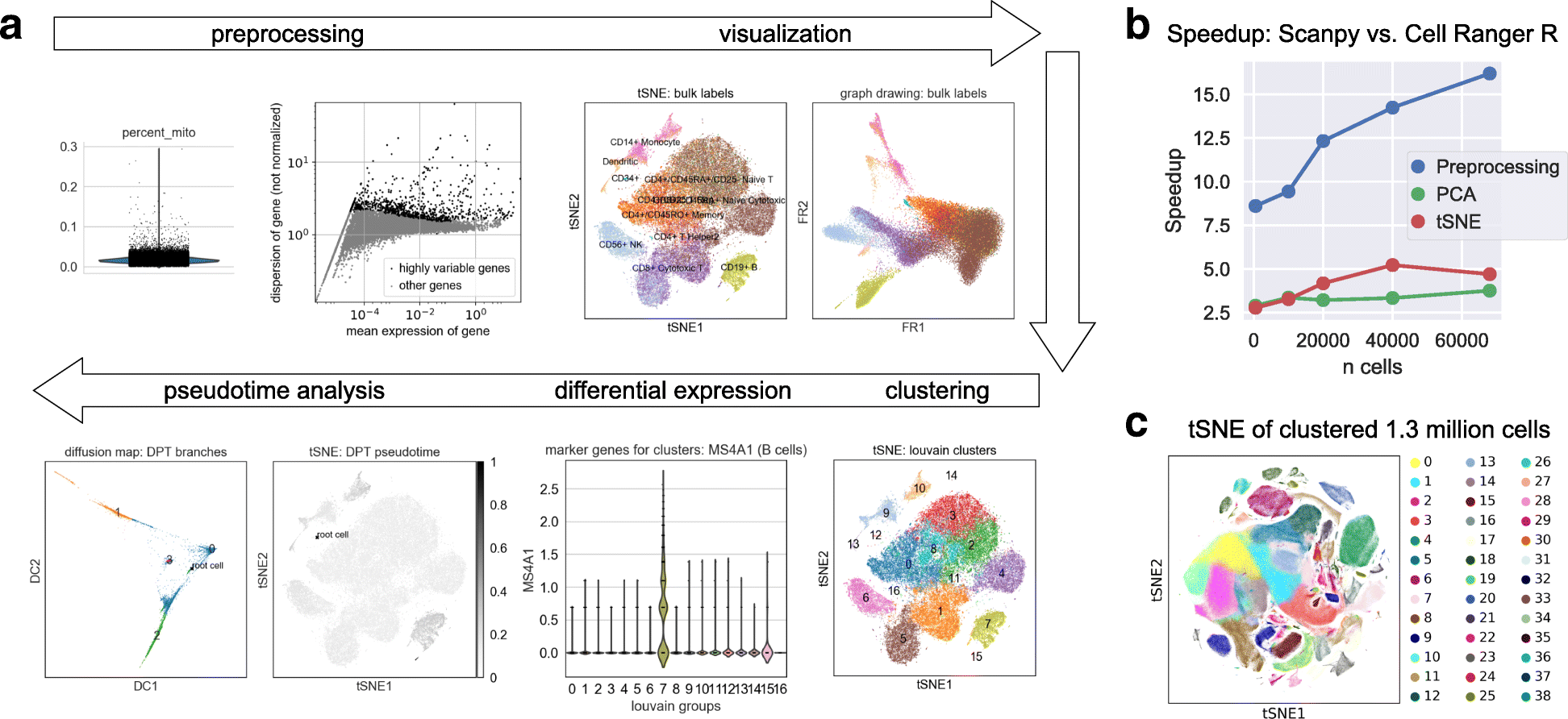
SCANPY is a scalable toolkit for analyzing single-cell gene expression data. It includes methods for preprocessing, visualization, clustering, pseudotime and trajectory inference, differential expression testing, and simulation of gene regulatory networks. Its Python-based implementation efficiently deals with data sets of more than one million cells ( https://github.com/theislab/Scanpy ). Along with SCANPY, we present ANNDATA, a generic class for handling annotated data matrices ( https://github.com/theislab/anndata ).
Keywords: Bioinformatics; Clustering; Differential expression testing; Graph analysis; Machine learning; Pseudotemporal ordering; Scalability; Single-cell transcriptomics; Trajectory inference; Visualization.
Conflict of interest statement
Ethics approval and consent to participate
Ethics approval was not applicable for this study.
Competing interests
None of the authors declare competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

